library(rpact)
packageVersion("rpact") # version should be version 3.0 or laterHow to Create One- and Multi-Arm Simulation Result Plots with rpact
Utilities
Power simulation
Multi-arm
Preparation
First, load the rpact package
[1] '4.3.0'Simulation results base
Simulation results base - means
maxNumberOfSubjects <- 90
informationRates <- c(0.2, 0.5, 1)
plannedSubjects <- round(informationRates * maxNumberOfSubjects)
getDesignInverseNormal(
futilityBounds = c(-0.5, 0.5),
informationRates = informationRates
) |>
getSimulationMeans(
groups = 2,
meanRatio = TRUE,
thetaH0 = 0.4,
plannedSubjects = plannedSubjects,
maxNumberOfIterations = 500,
allocationRatioPlanned = 3,
stDev = 1.5,
seed = 1234567890
) |>
plot(type = "all", grid = 0)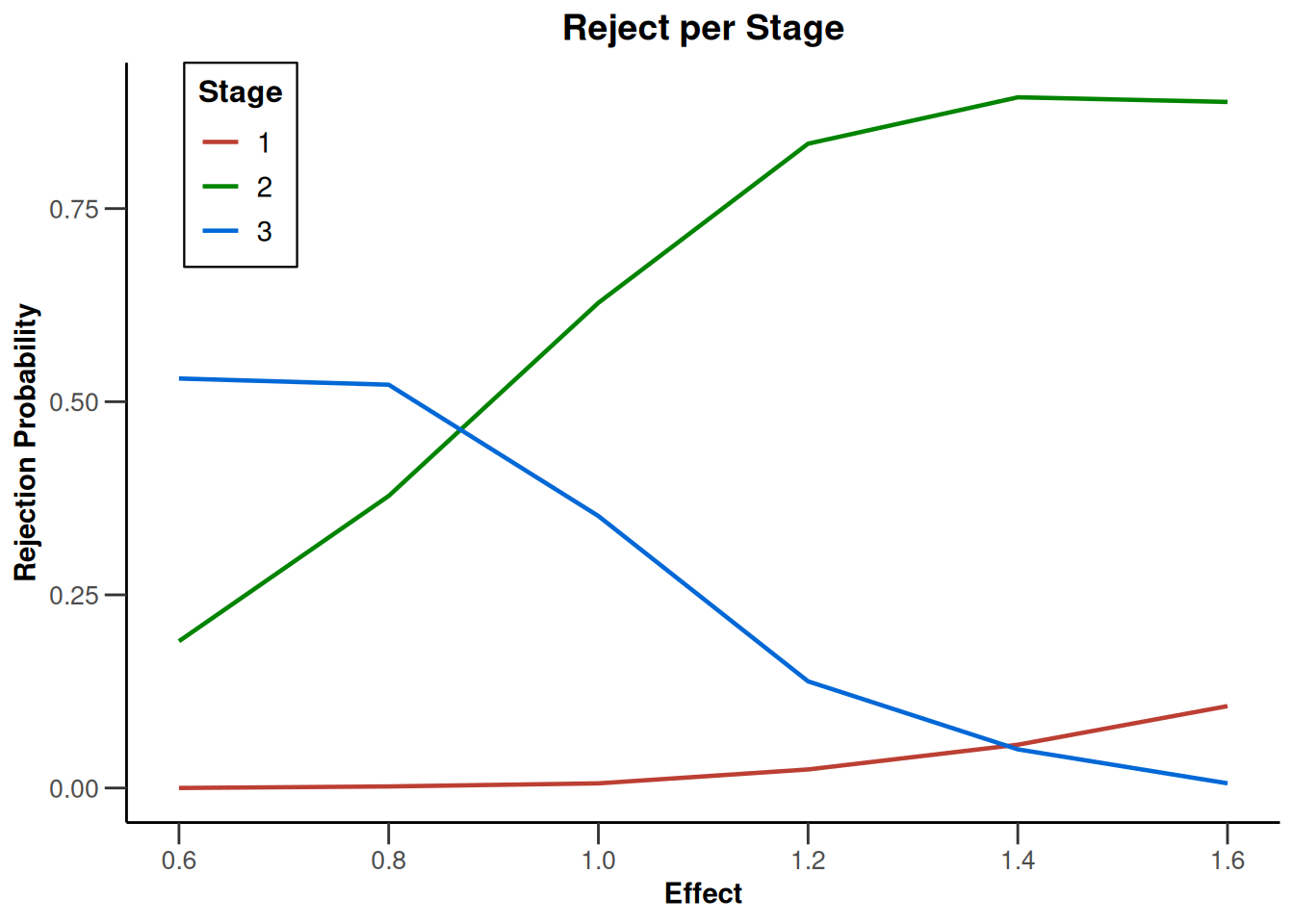
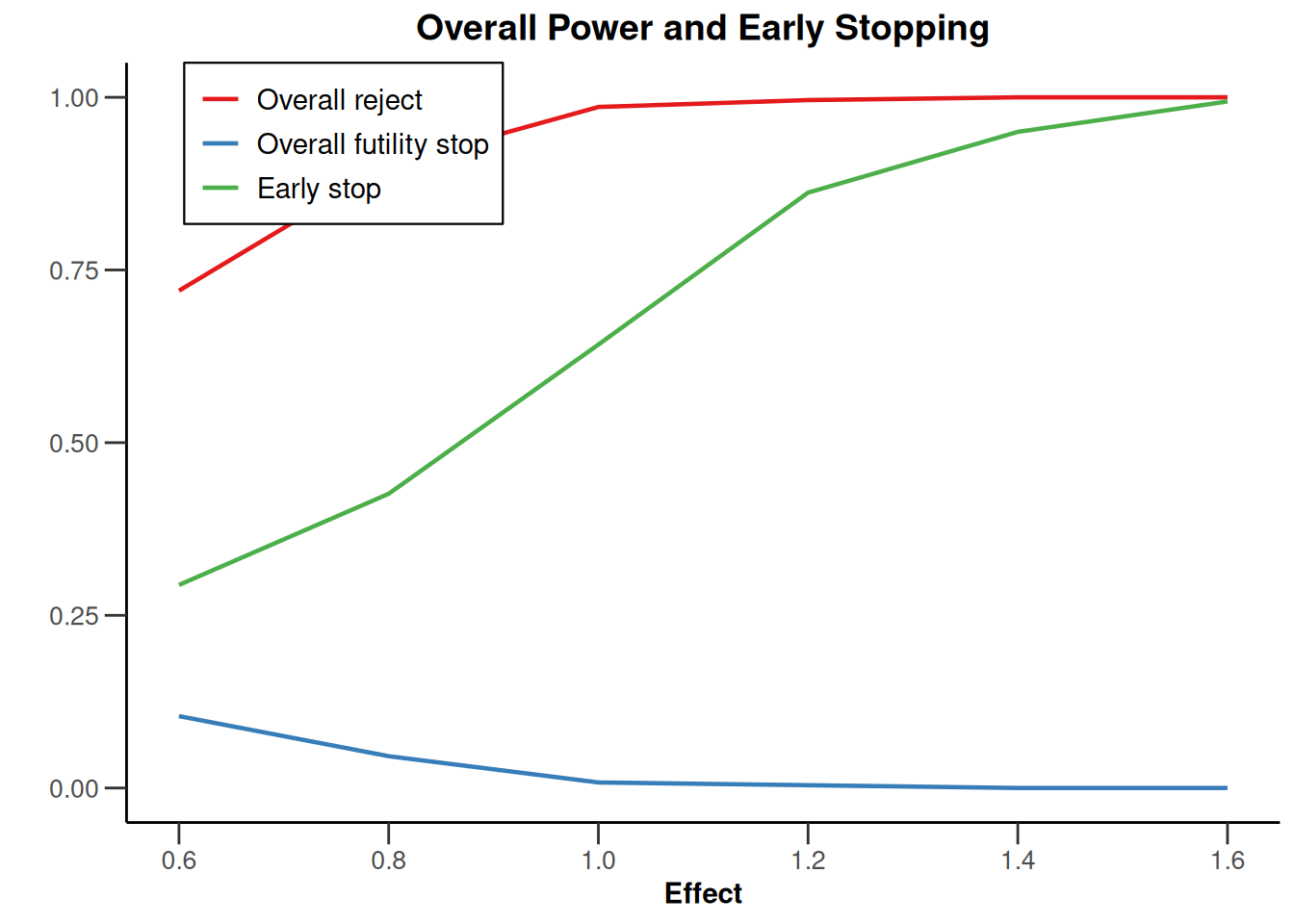
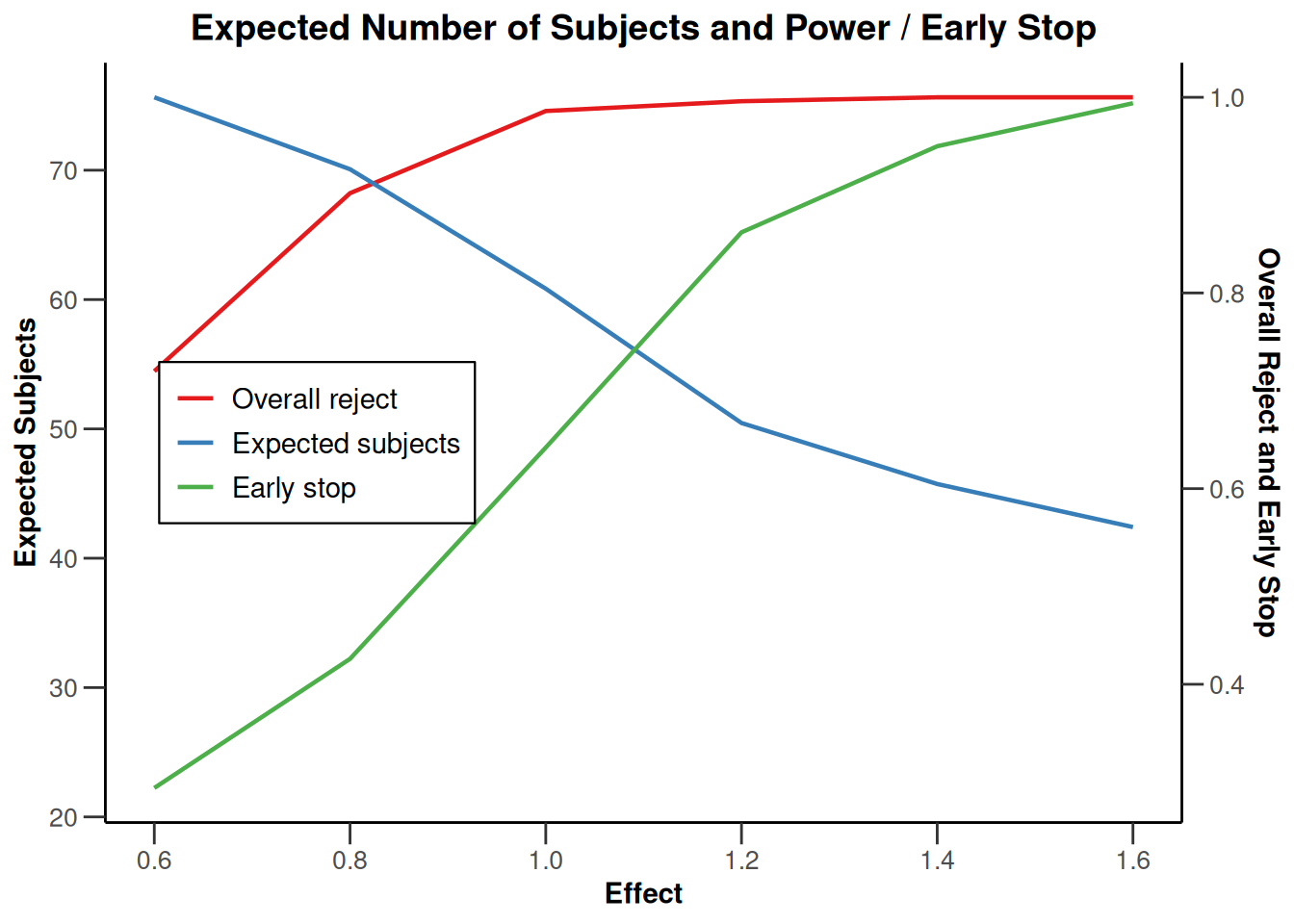
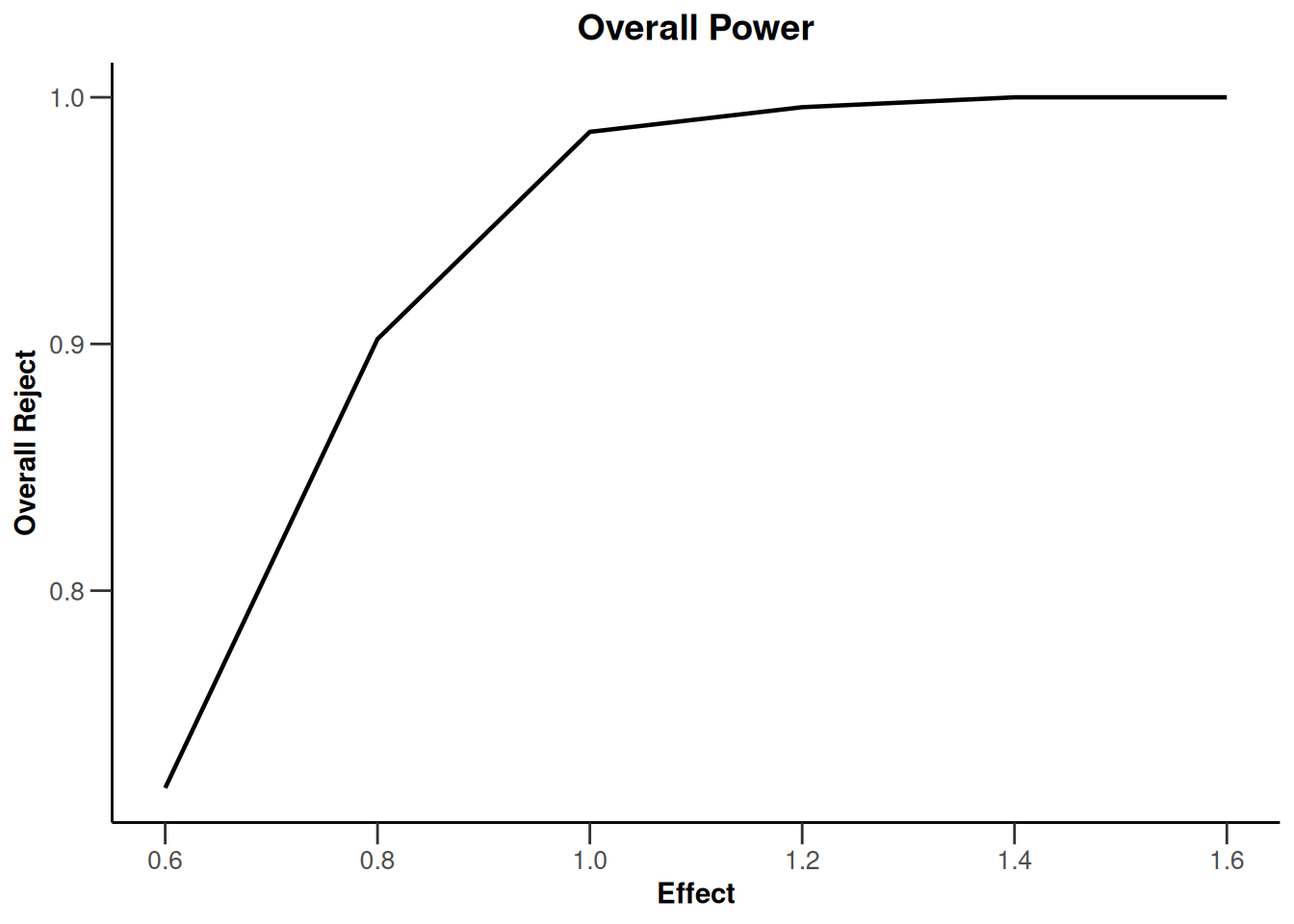
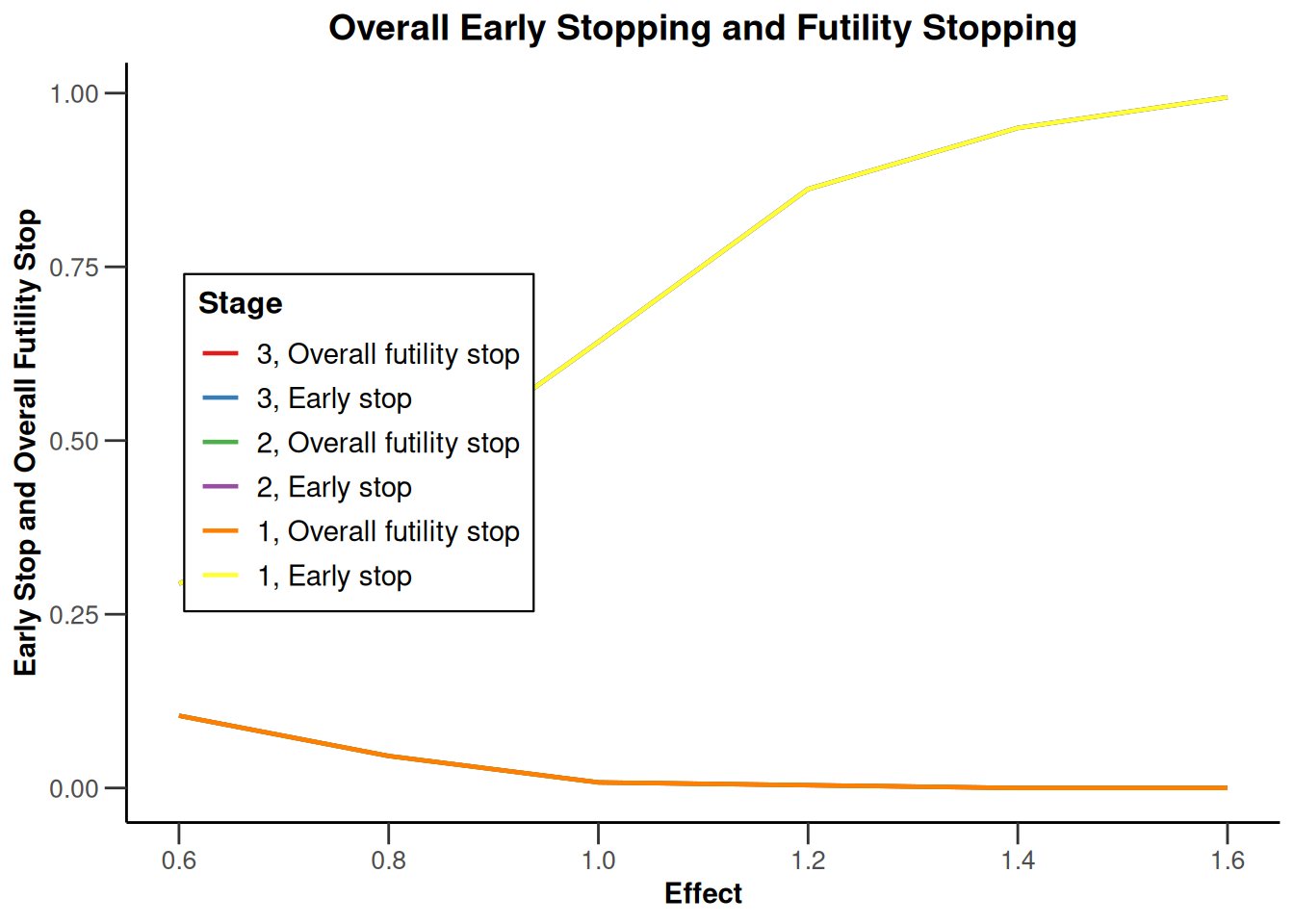
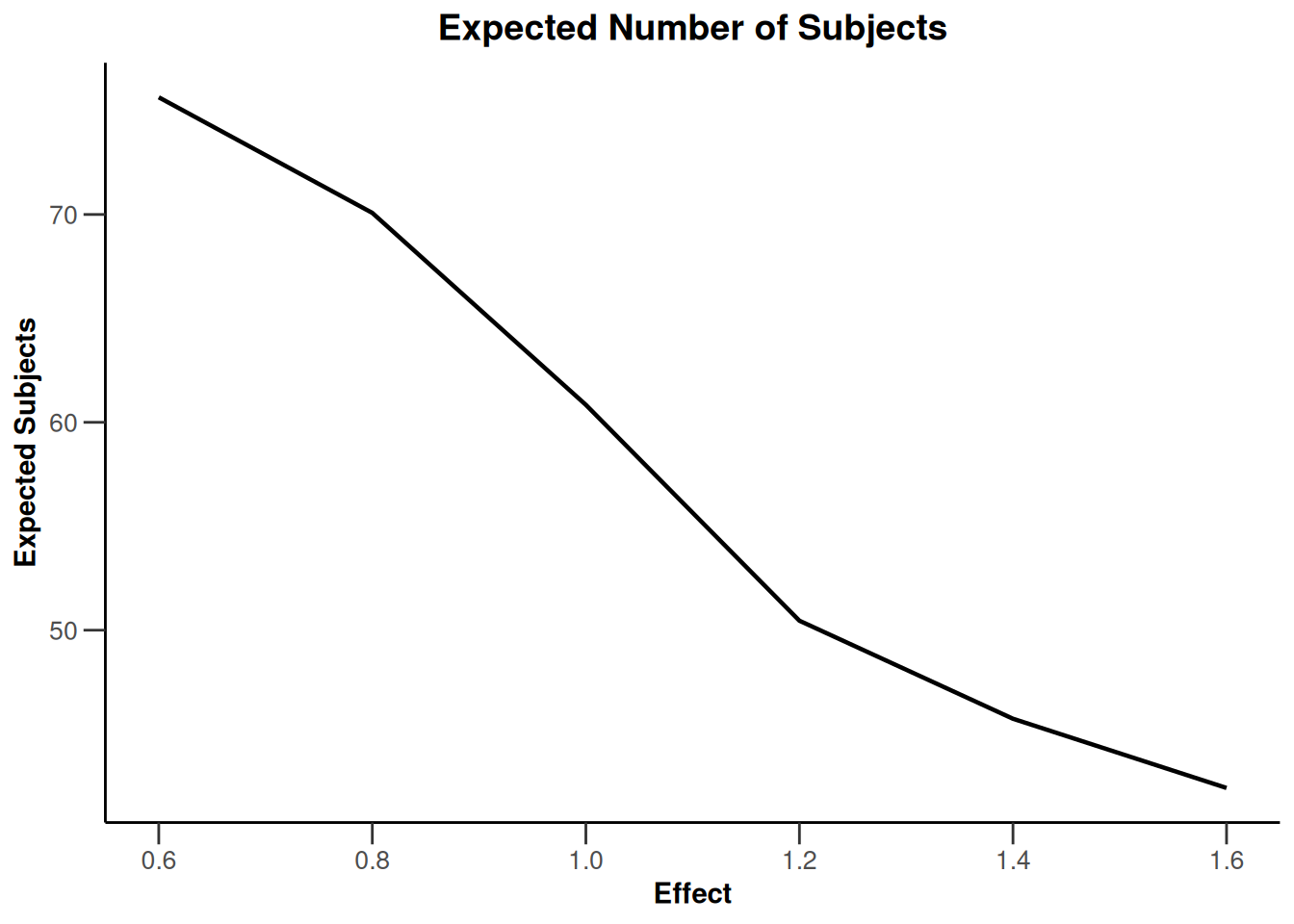
Simulation results base - rates
maxNumberOfSubjects <- 90
informationRates <- (1:3) / 3
plannedSubjects <- round(informationRates * maxNumberOfSubjects)
getDesignInverseNormal(
futilityBounds = c(-0.5, 0.5),
informationRates = informationRates
) |>
getSimulationRates(
groups = 2,
riskRatio = TRUE,
thetaH0 = 0.8,
plannedSubjects = plannedSubjects,
maxNumberOfIterations = 500,
allocationRatioPlanned = 3,
seed = 1234567890
) |>
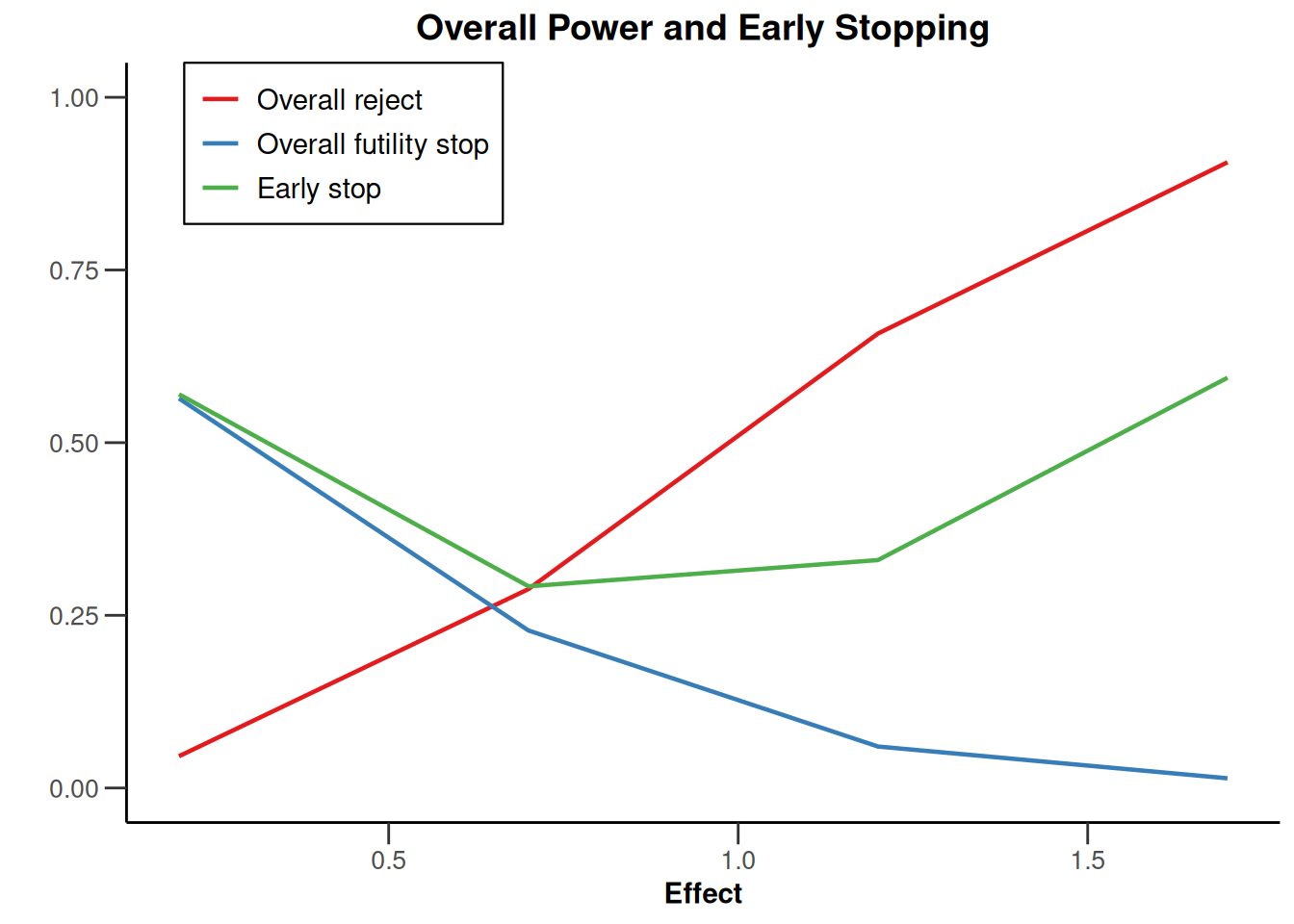
plot(type = "all", grid = 0)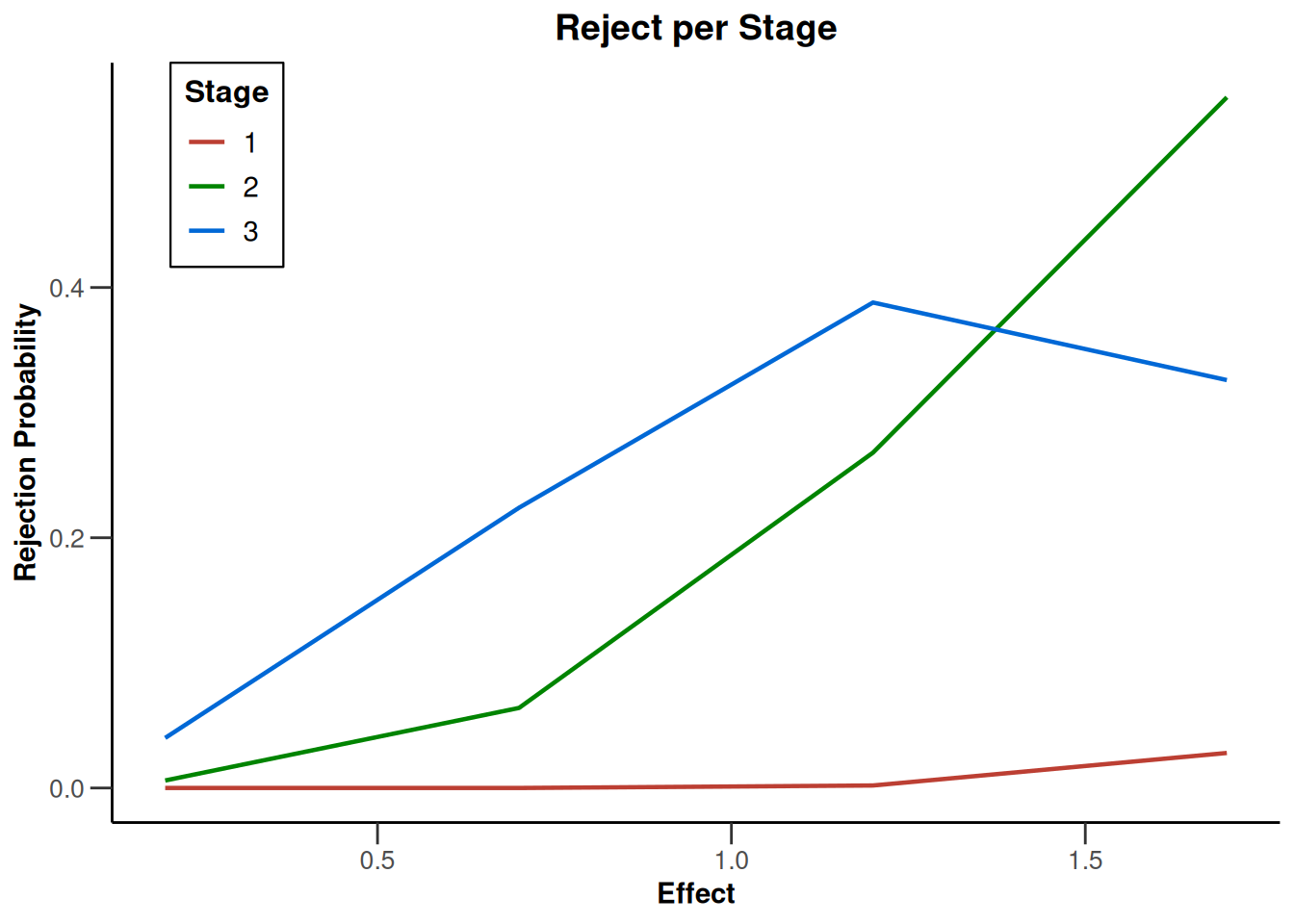
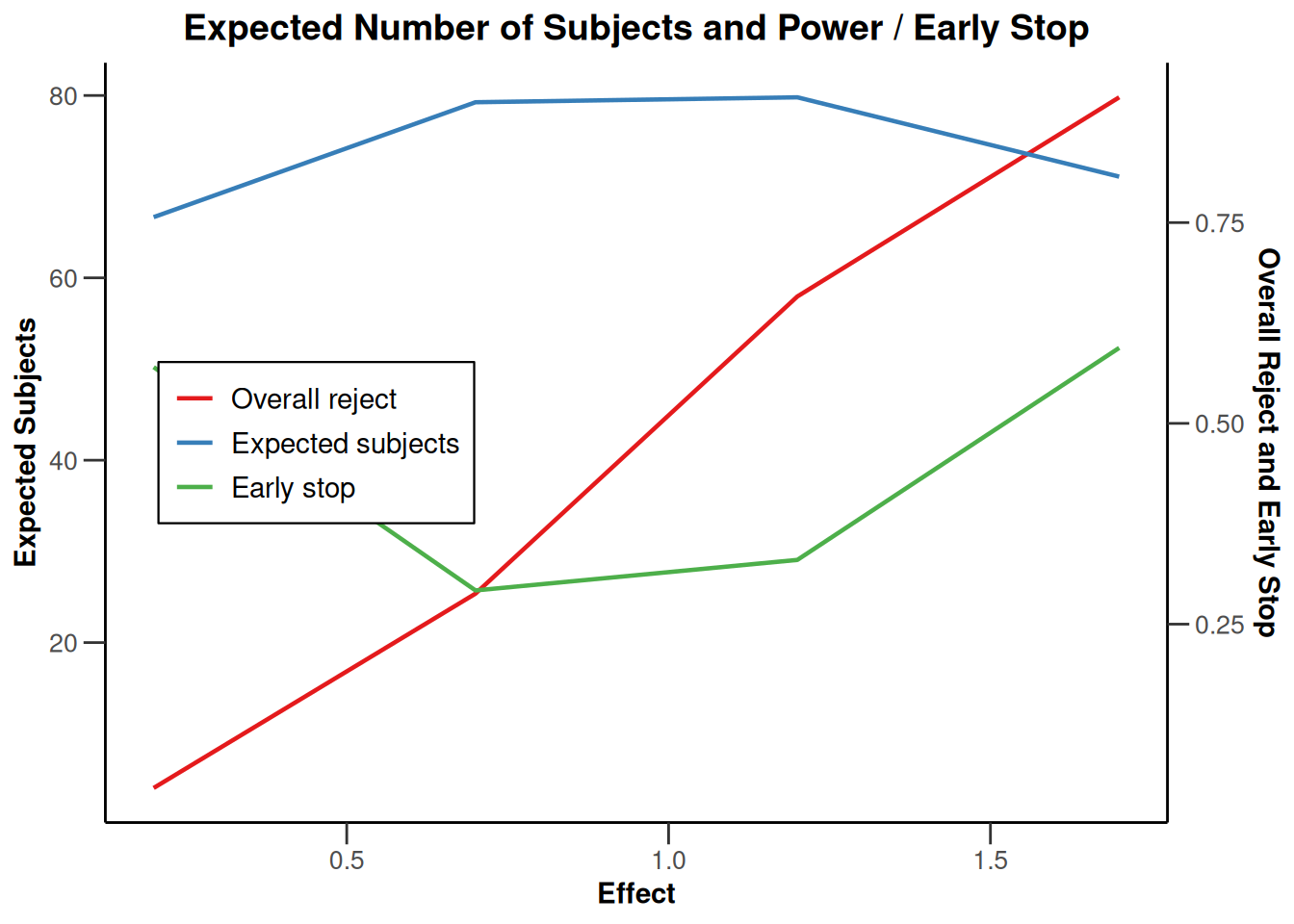
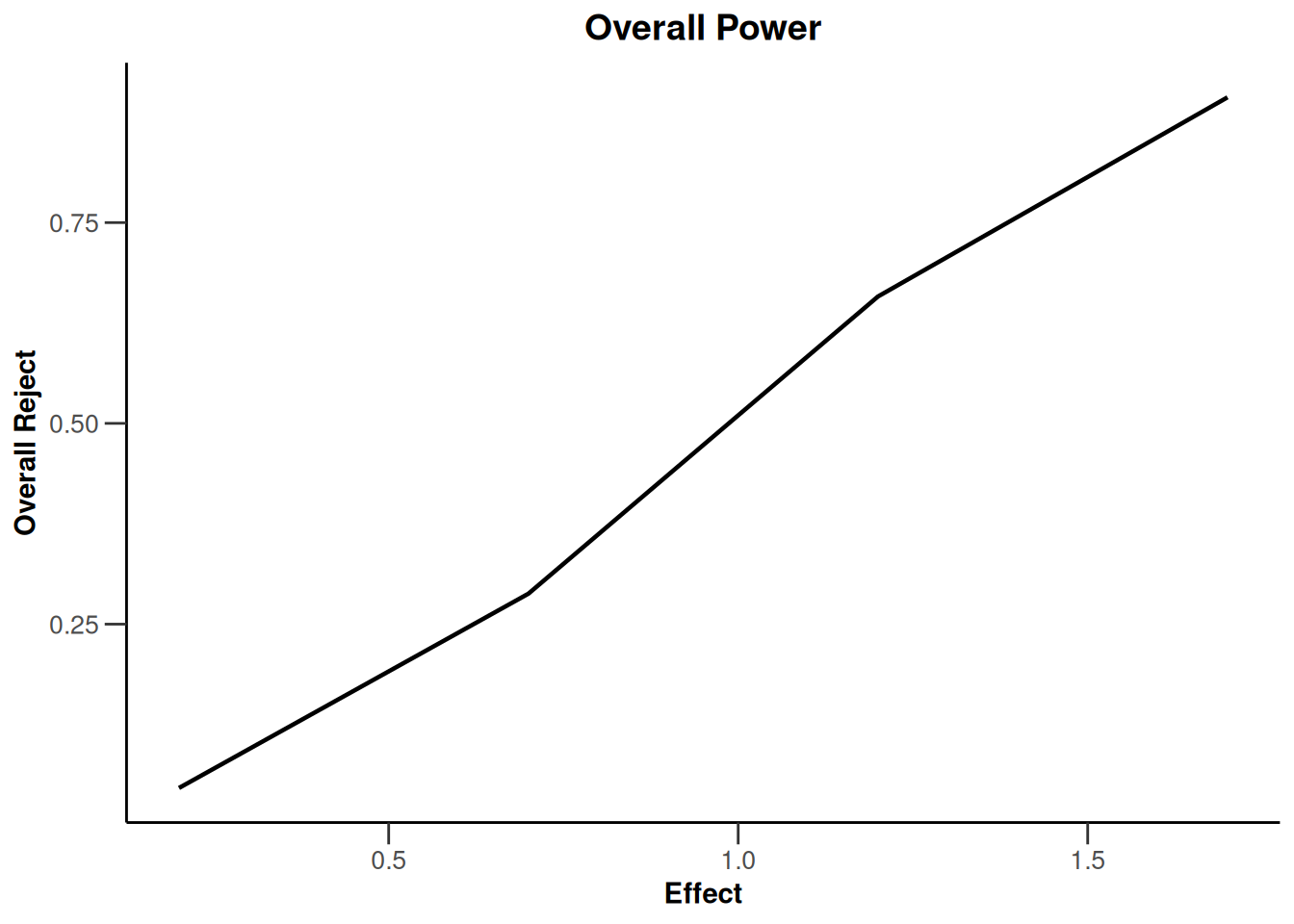
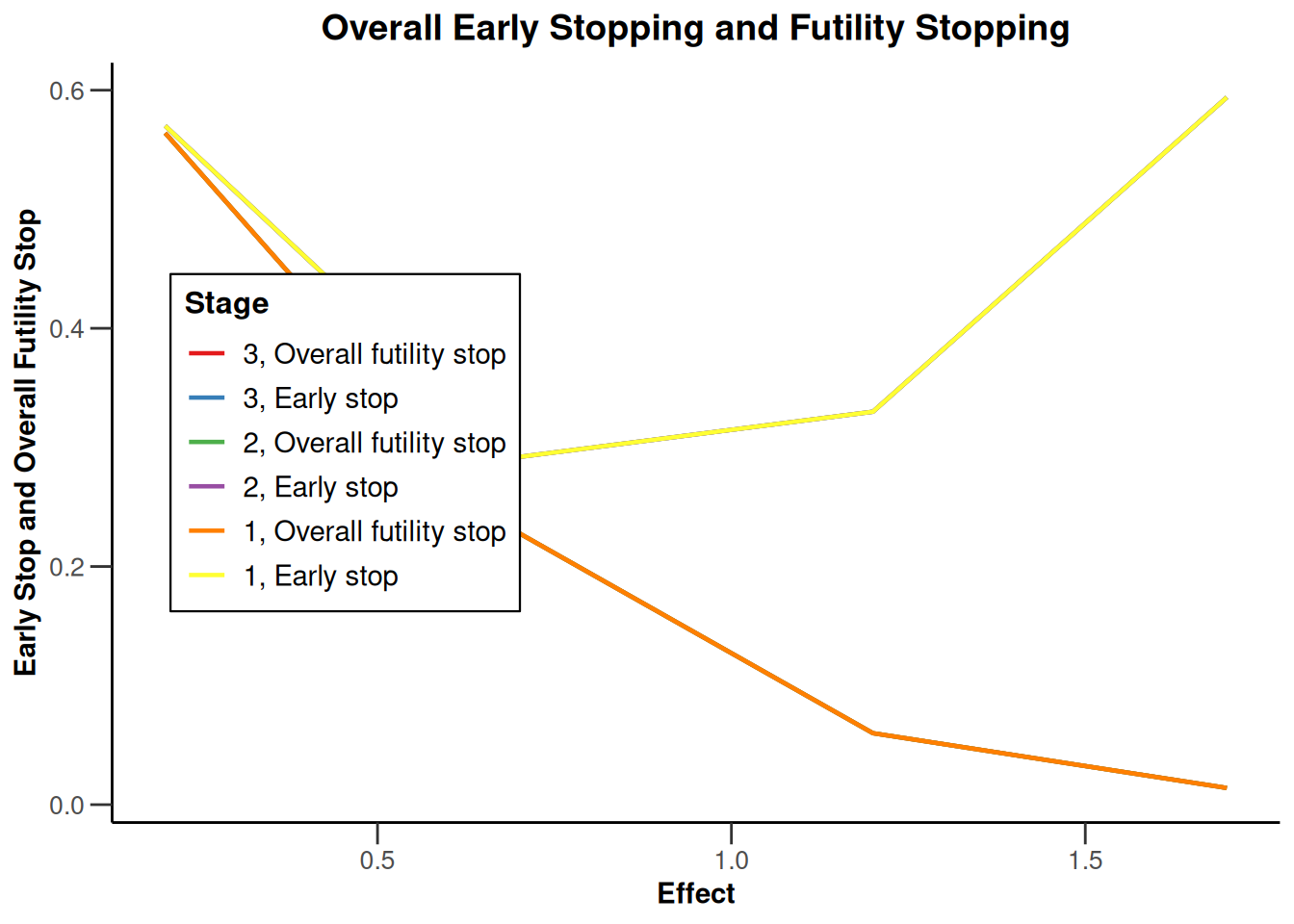
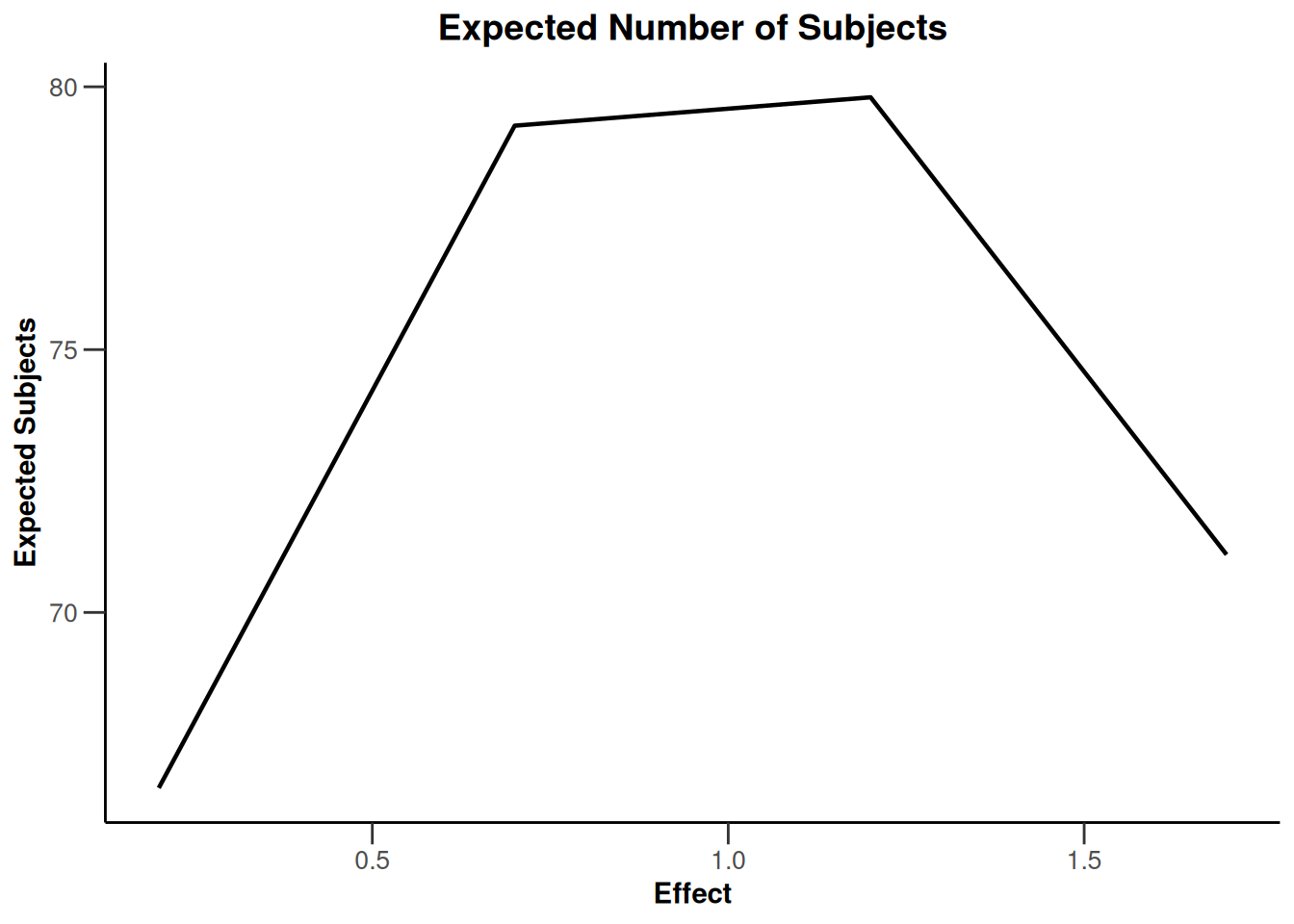
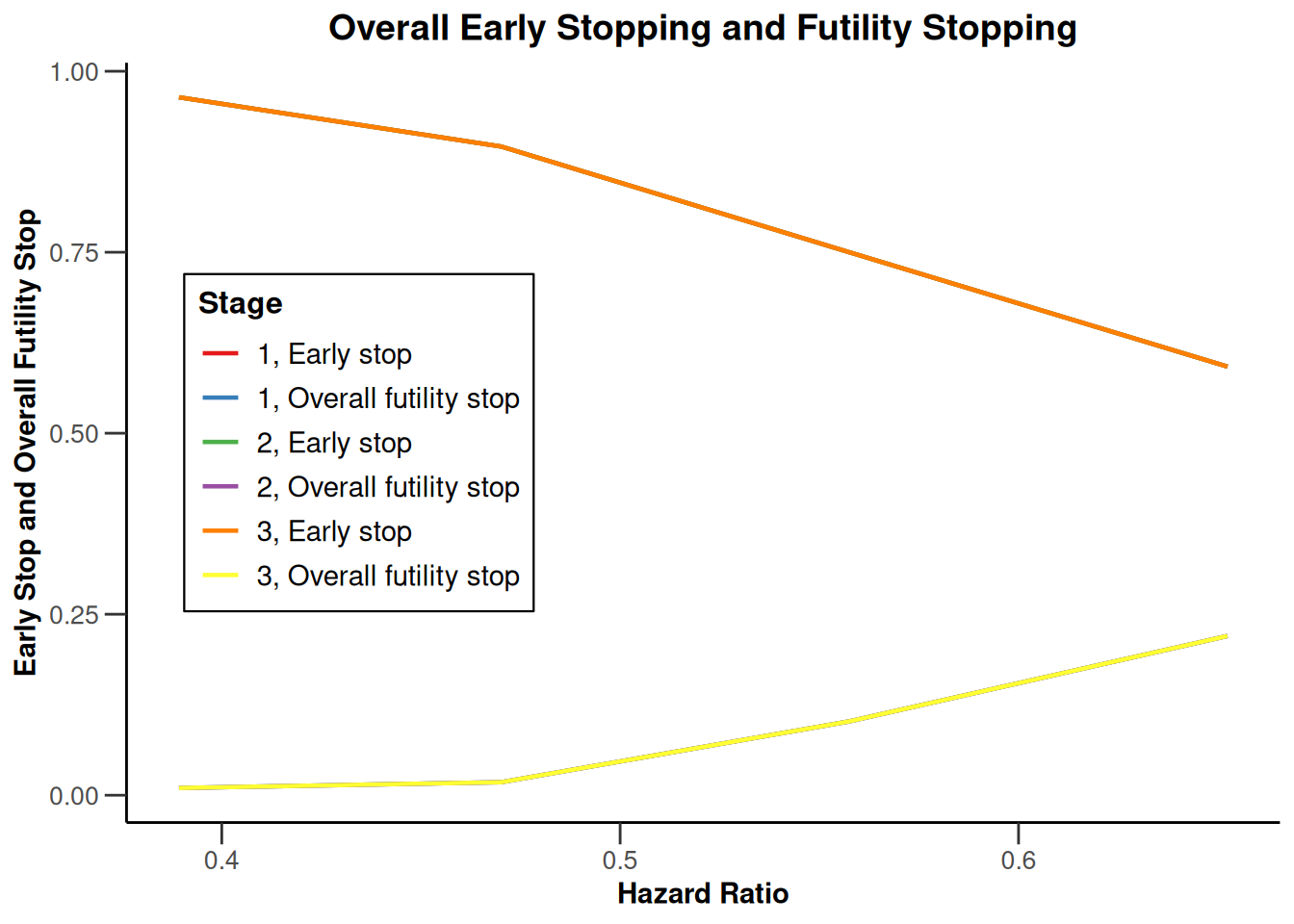
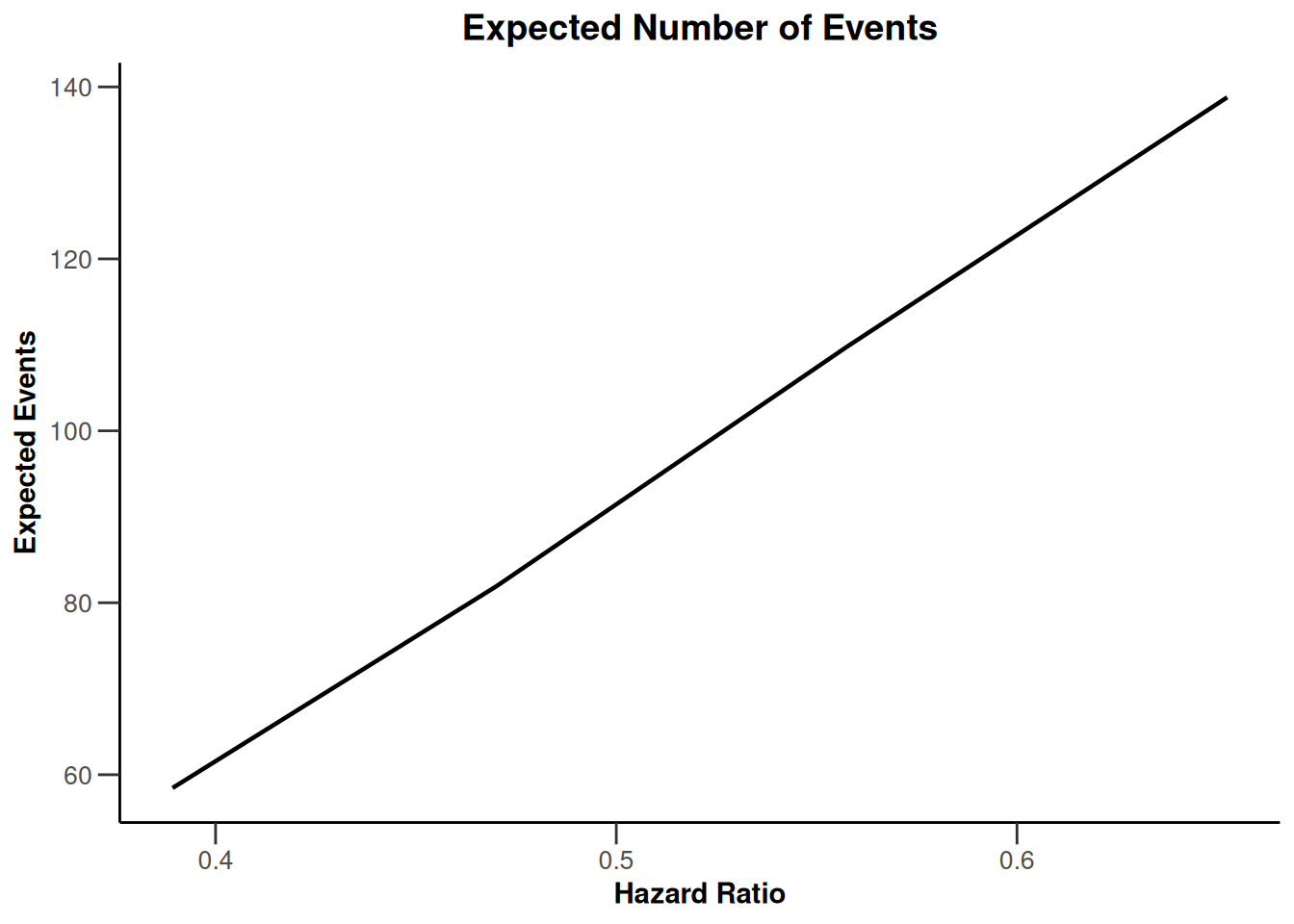
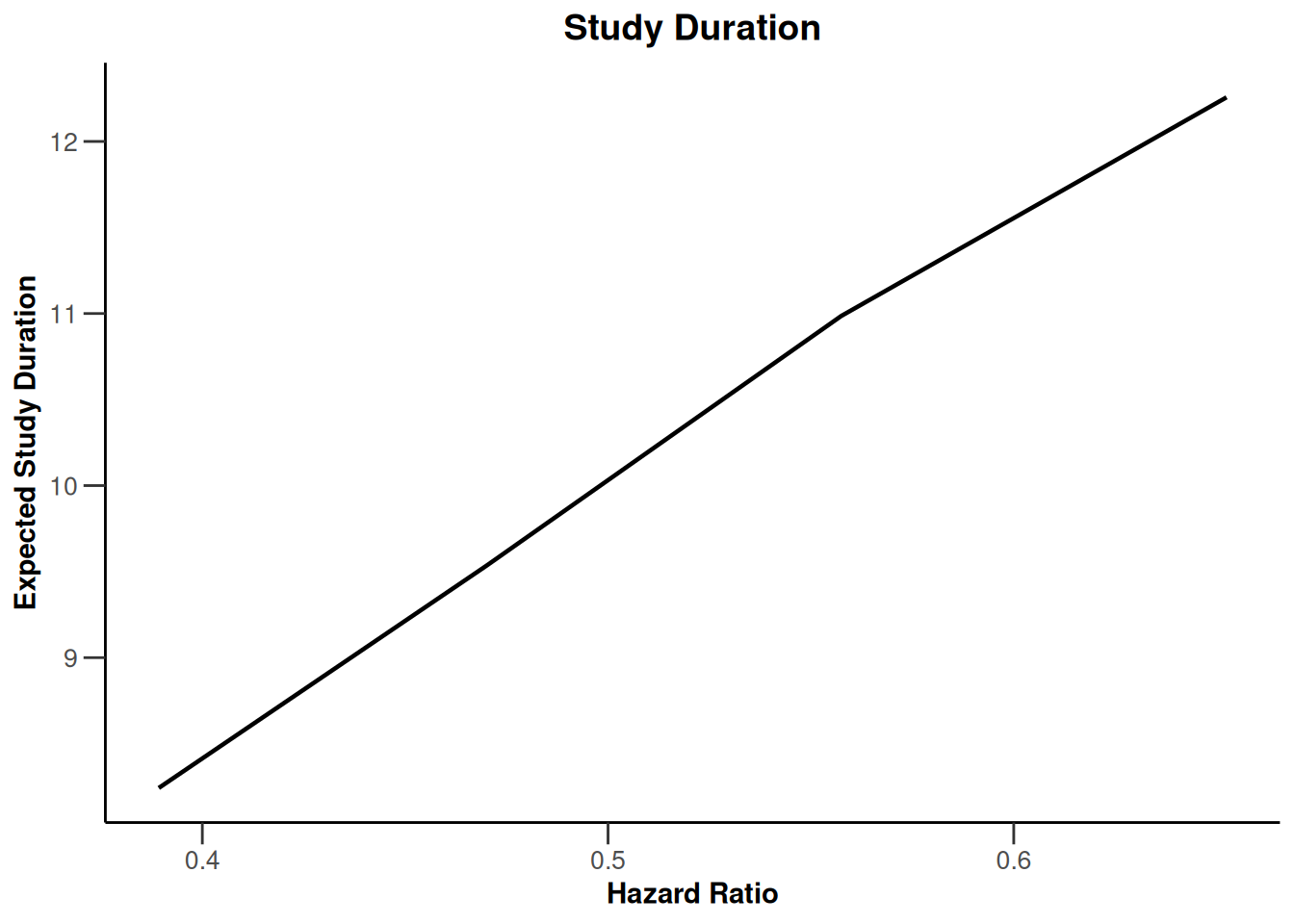
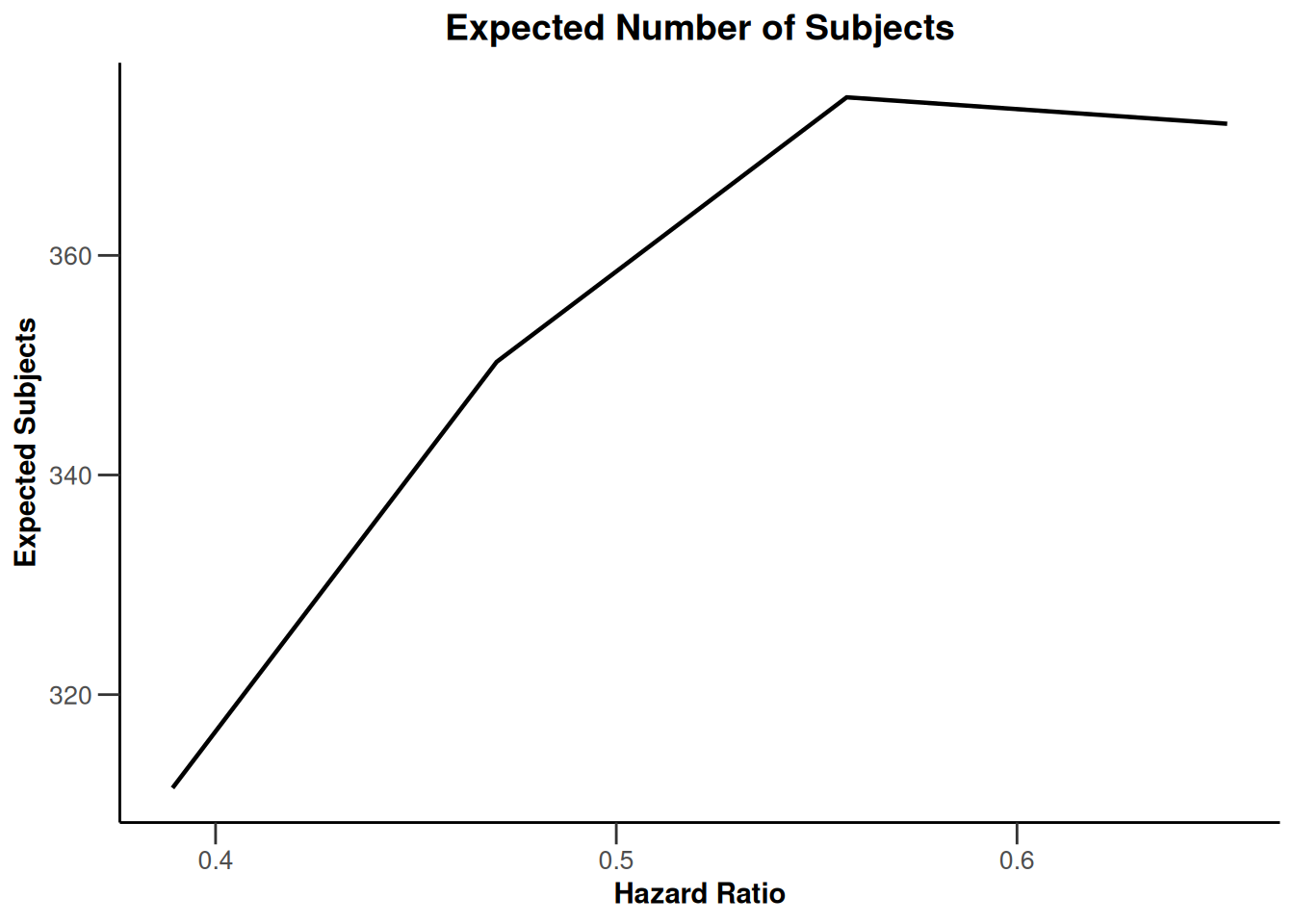
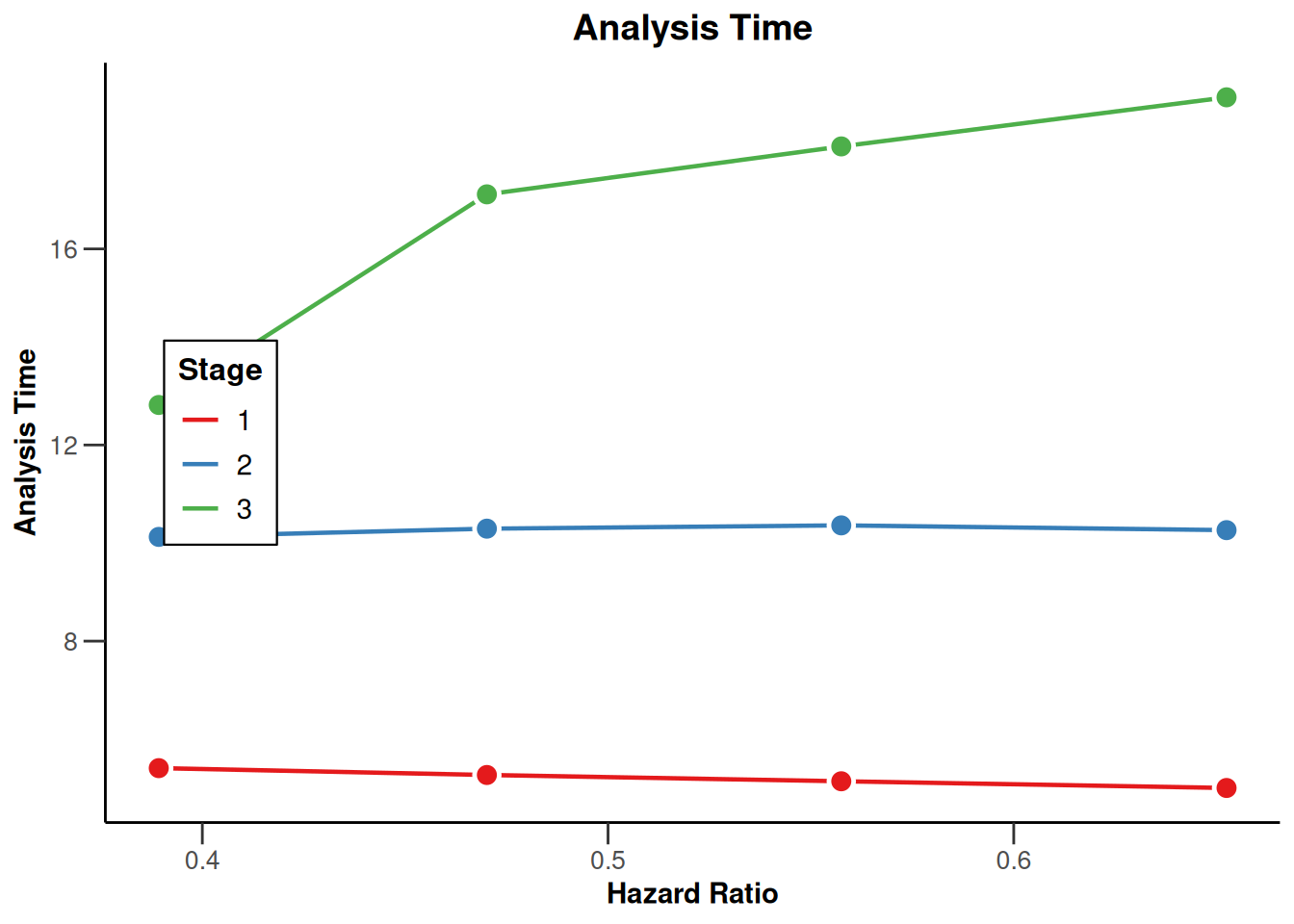
Simulation results base - survival
kMax <- 3
getDesignFisher(
kMax = kMax,
alpha0Vec = c(0.5, 0.5)
) |>
getSimulationSurvival(
pi2 = 0.6,
pi1 = seq(0.3, 0.45, 0.05),
directionUpper = FALSE,
maxNumberOfSubjects = 500,
plannedEvents = (1:kMax) * 20,
allocation1 = 1,
allocation2 = 1,
accrualTime = c(0, 3, 6, 12),
accrualIntensity = c(0.1, 0.2, 0.2),
dropoutRate1 = 0,
dropoutRate2 = 0,
dropoutTime = 12,
conditionalPower = 0.8,
minNumberOfEventsPerStage = c(NA_real_, 10, 10),
maxNumberOfEventsPerStage = c(NA_real_, 100, 200),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)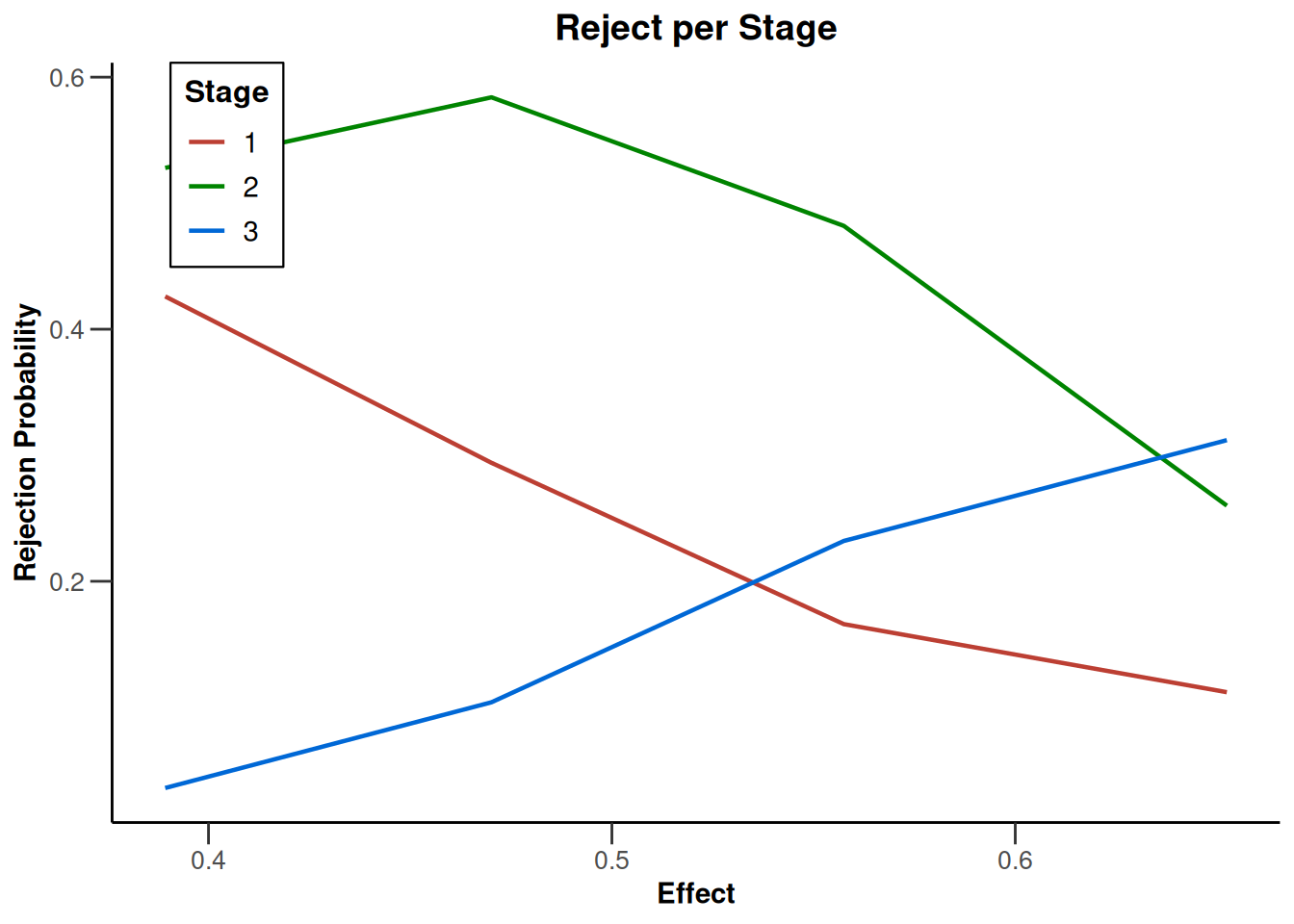
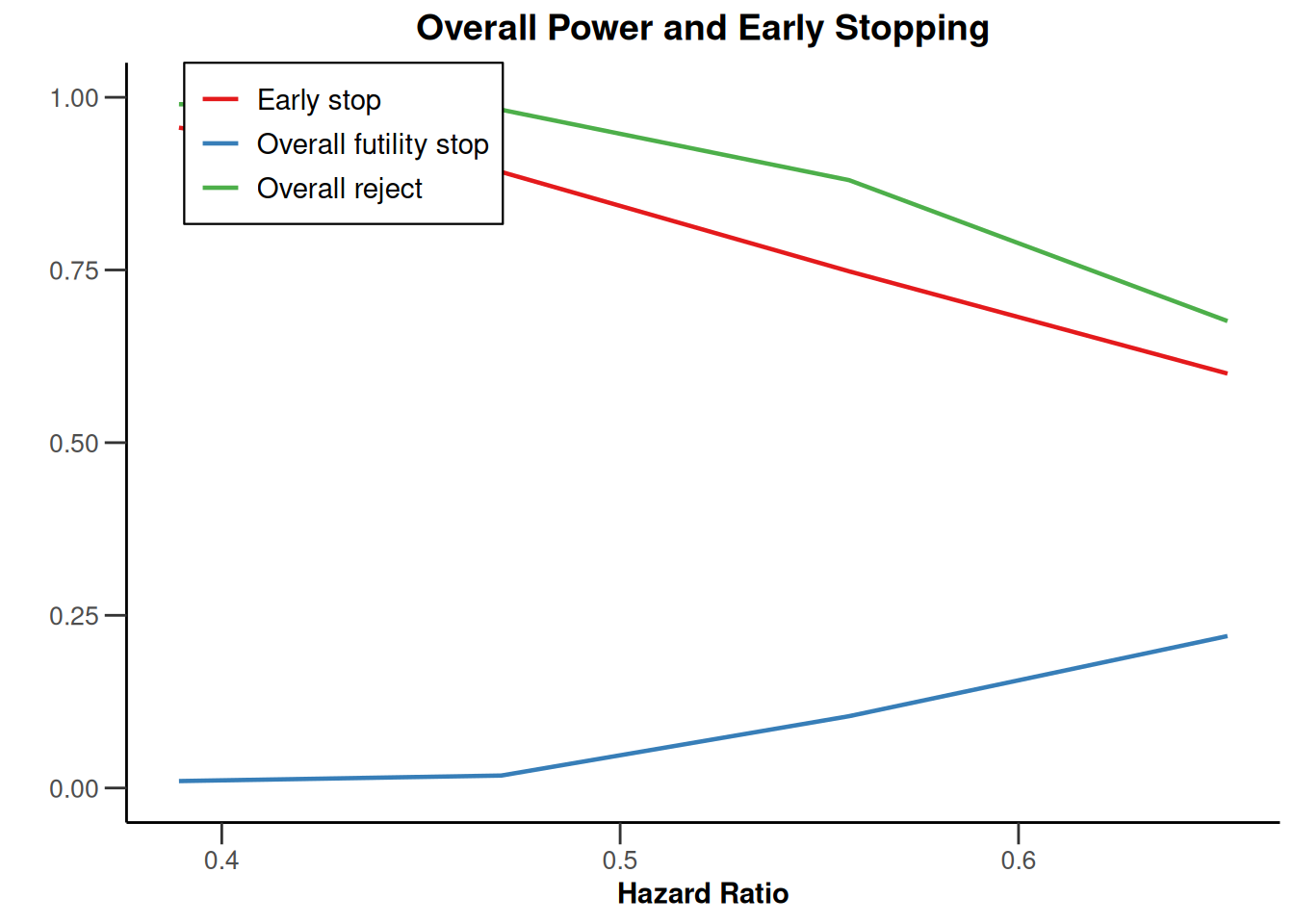
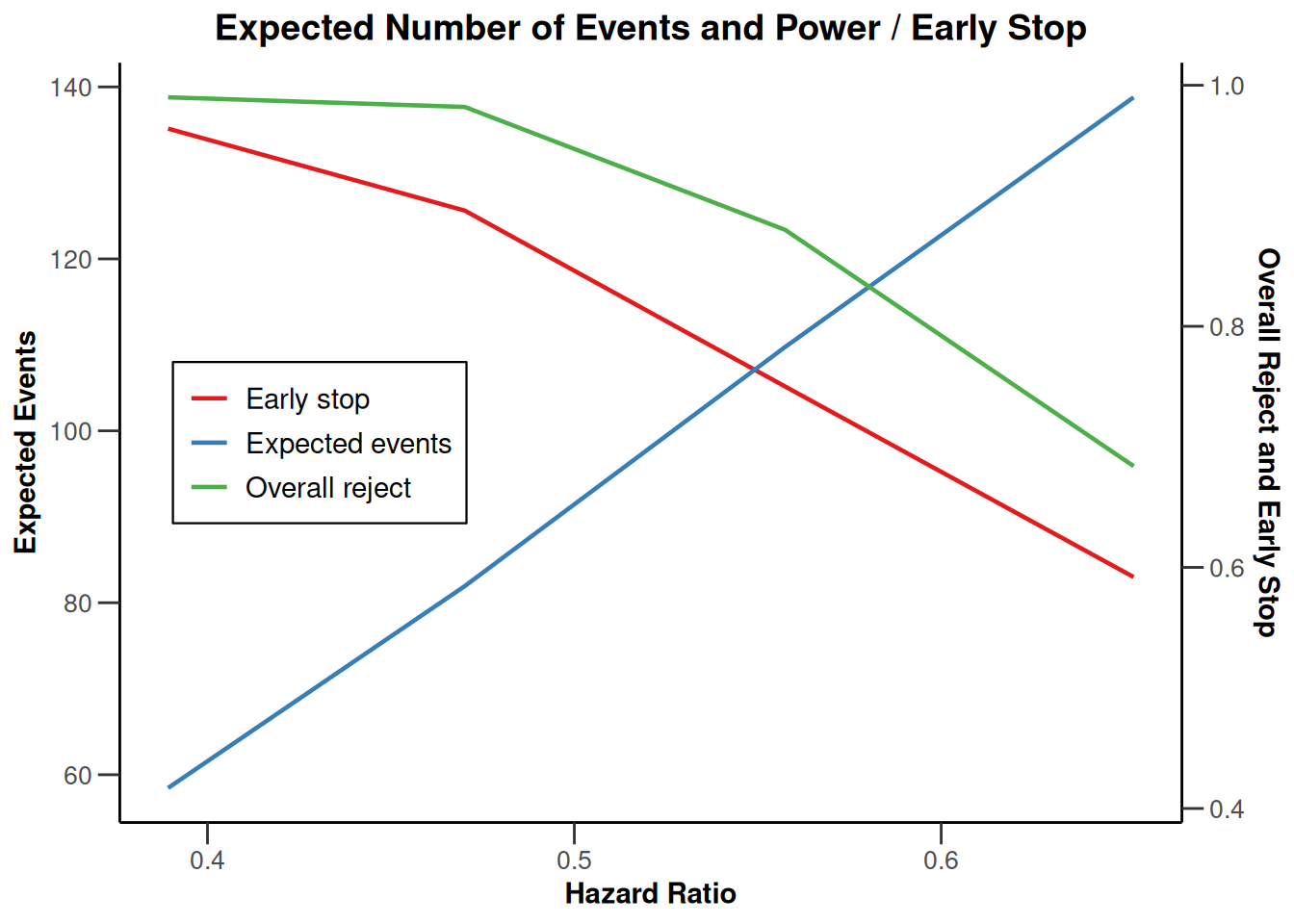
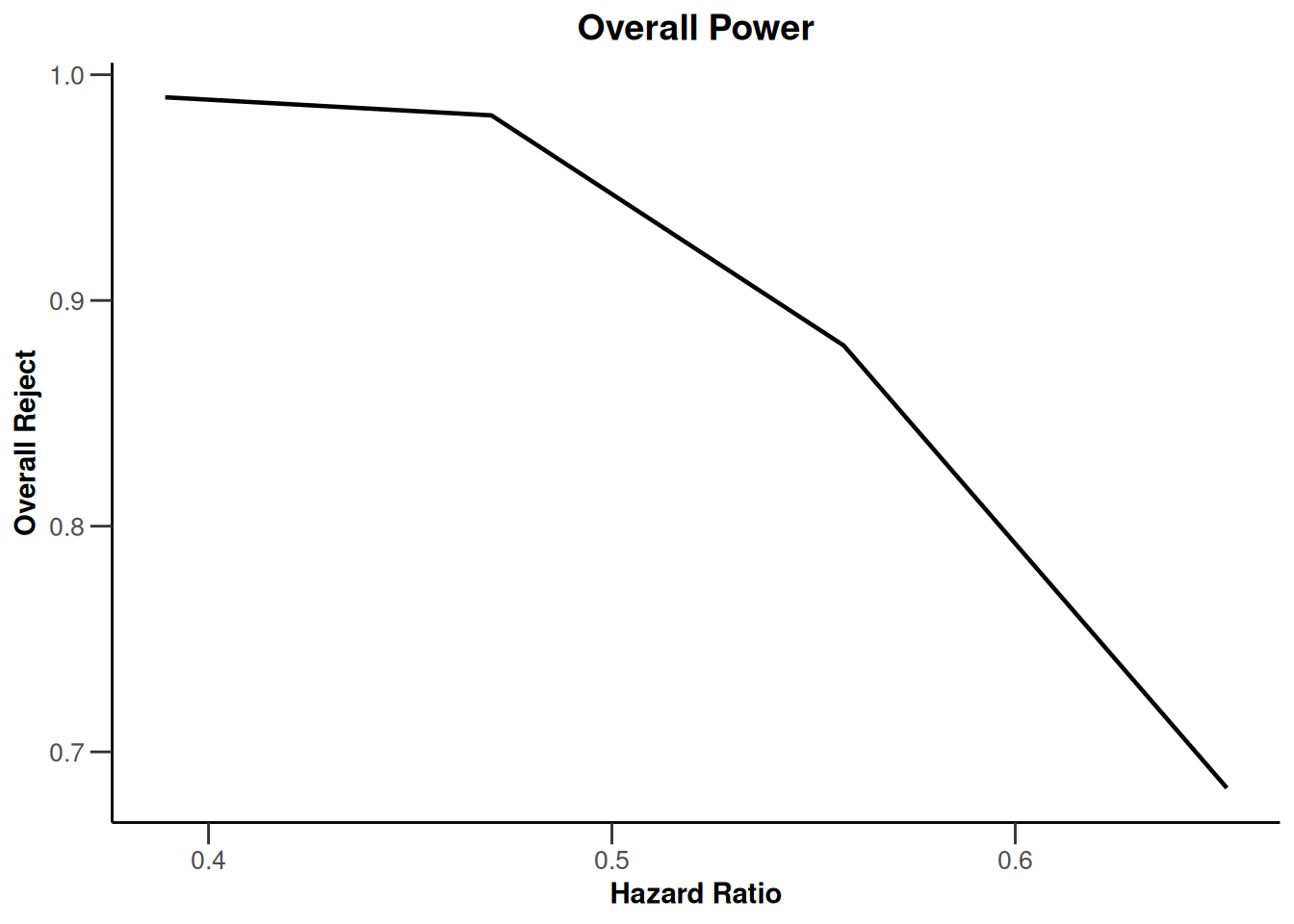
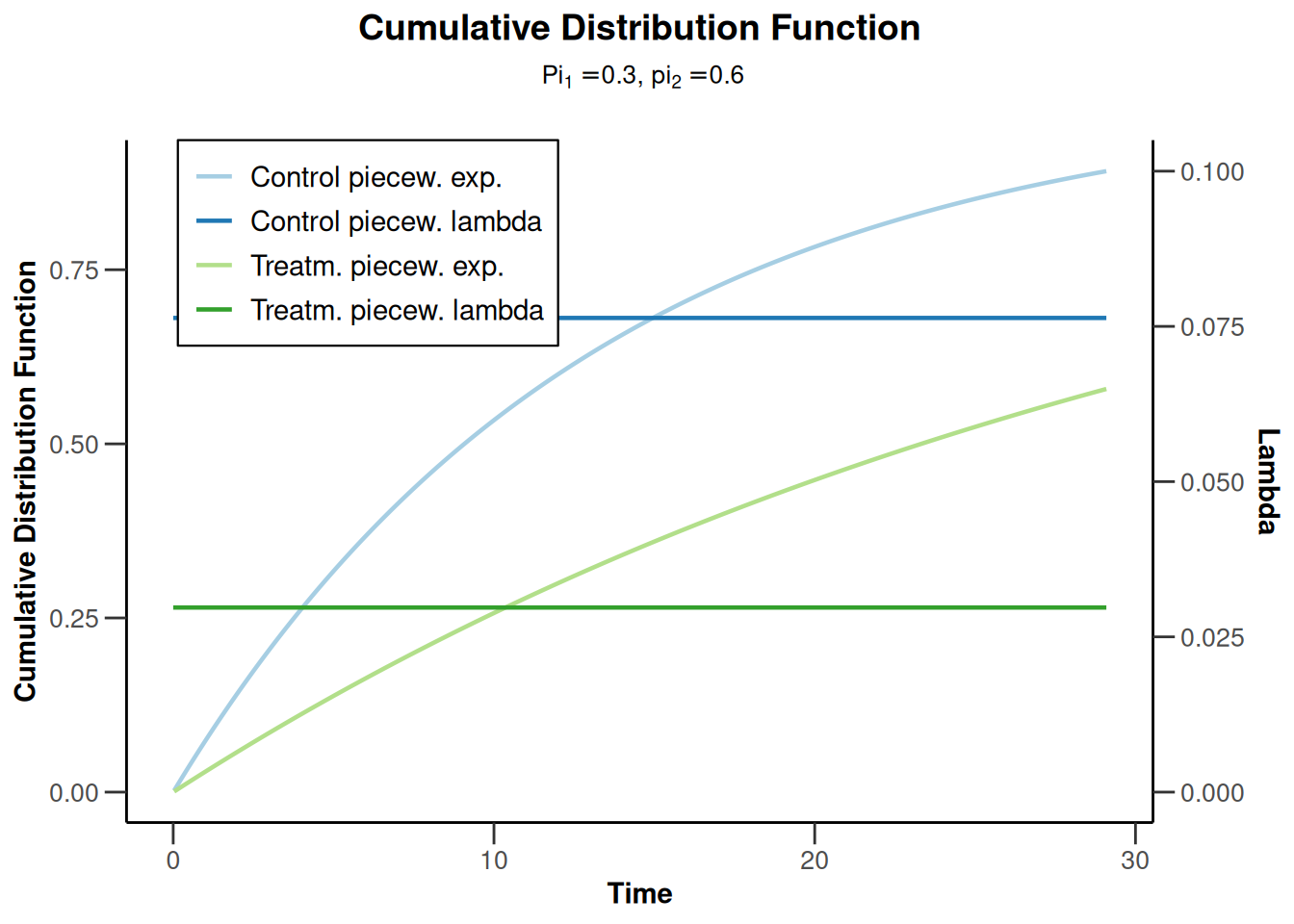
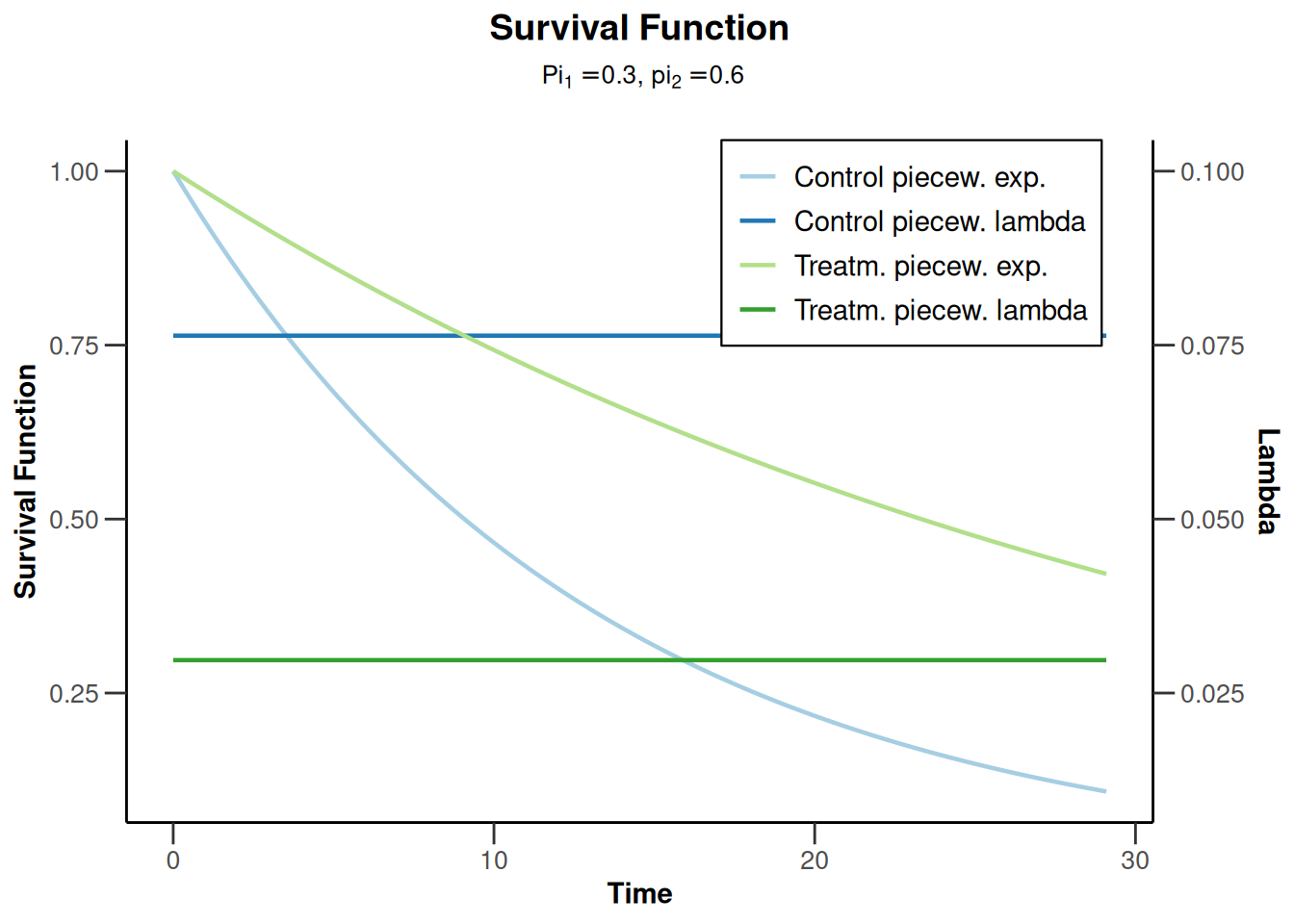
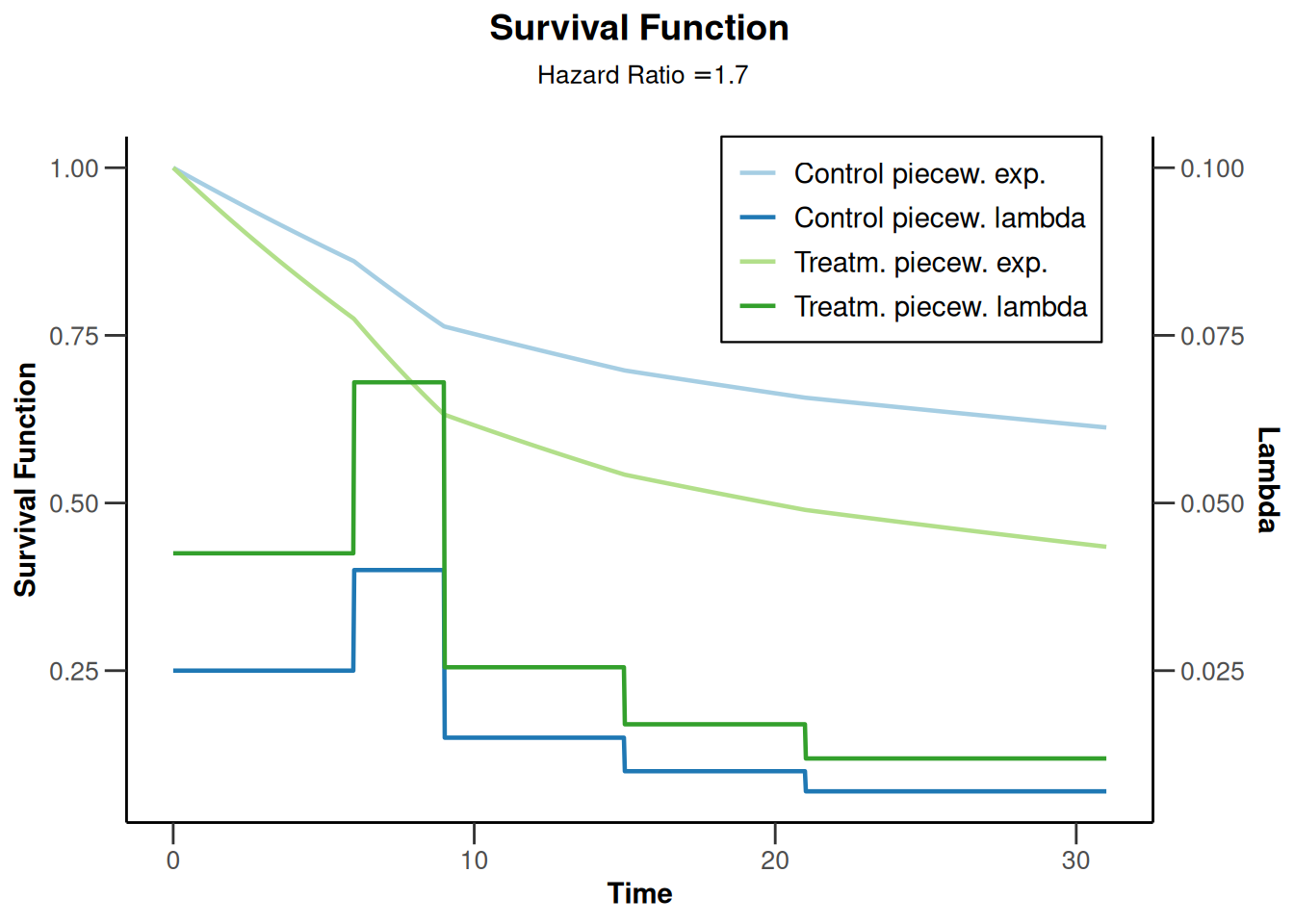
piecewiseSurvivalTime <- list(
"<6" = 0.025,
"6 - <9" = 0.04,
"9 - <15" = 0.015,
"15 - <21" = 0.01,
">=21" = 0.007
)
kMax <- 3
getDesignGroupSequential(
kMax = kMax,
typeOfDesign = "WT",
deltaWT = 0.25
) |>
getSimulationSurvival(
directionUpper = TRUE,
maxNumberOfSubjects = 500,
plannedEvents = (1:kMax) * 20,
allocation1 = 1,
allocation2 = 1,
accrualTime = c(0, 3, 6, 12),
piecewiseSurvivalTime = piecewiseSurvivalTime,
hazardRatio = 1.7,
accrualIntensity = c(0.1, 0.2, 0.2),
dropoutRate1 = 0,
dropoutRate2 = 0,
dropoutTime = 12,
conditionalPower = 0.8,
minNumberOfEventsPerStage = c(NA_real_, 10, 10),
maxNumberOfEventsPerStage = c(NA_real_, 100, 200),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)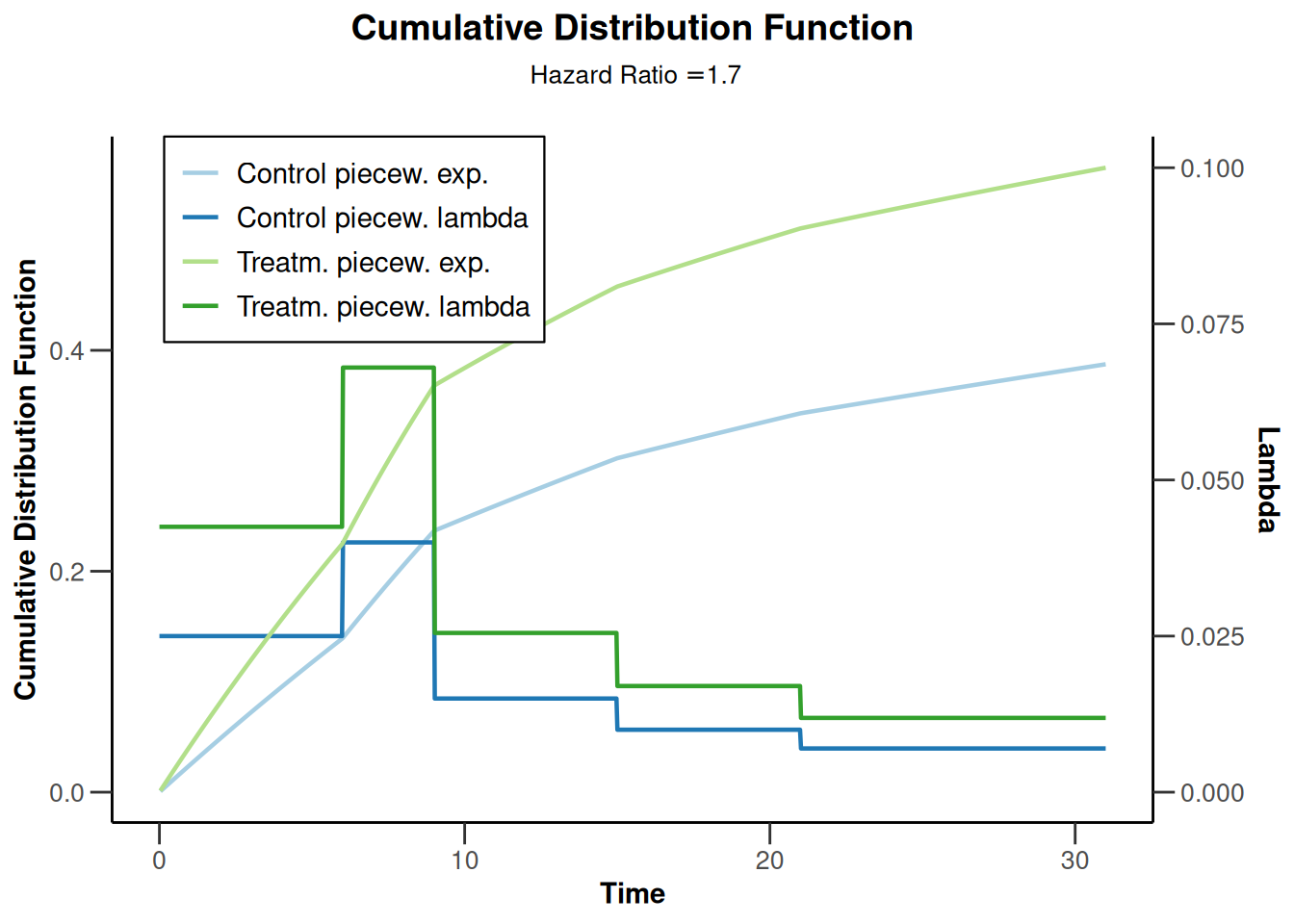
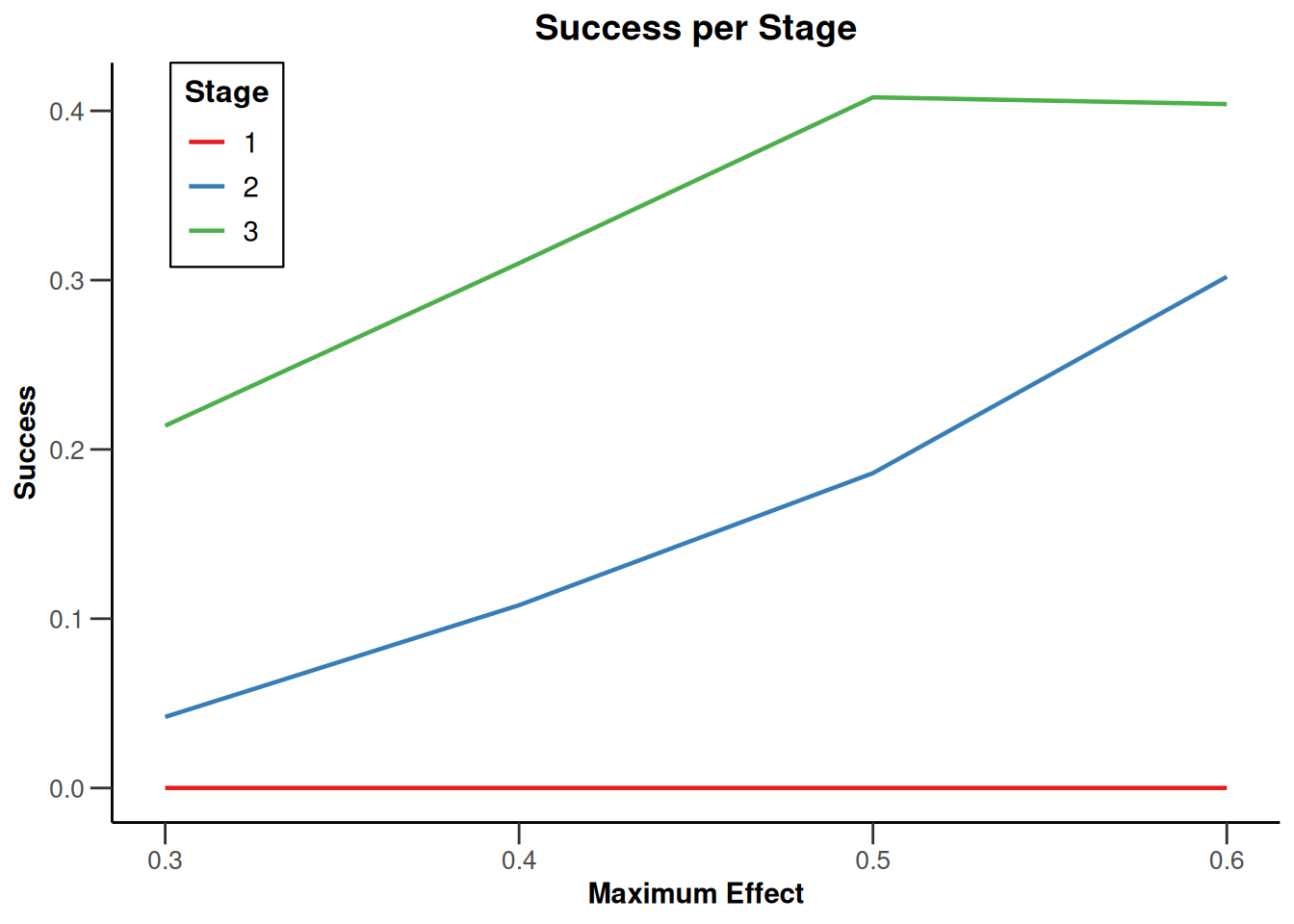
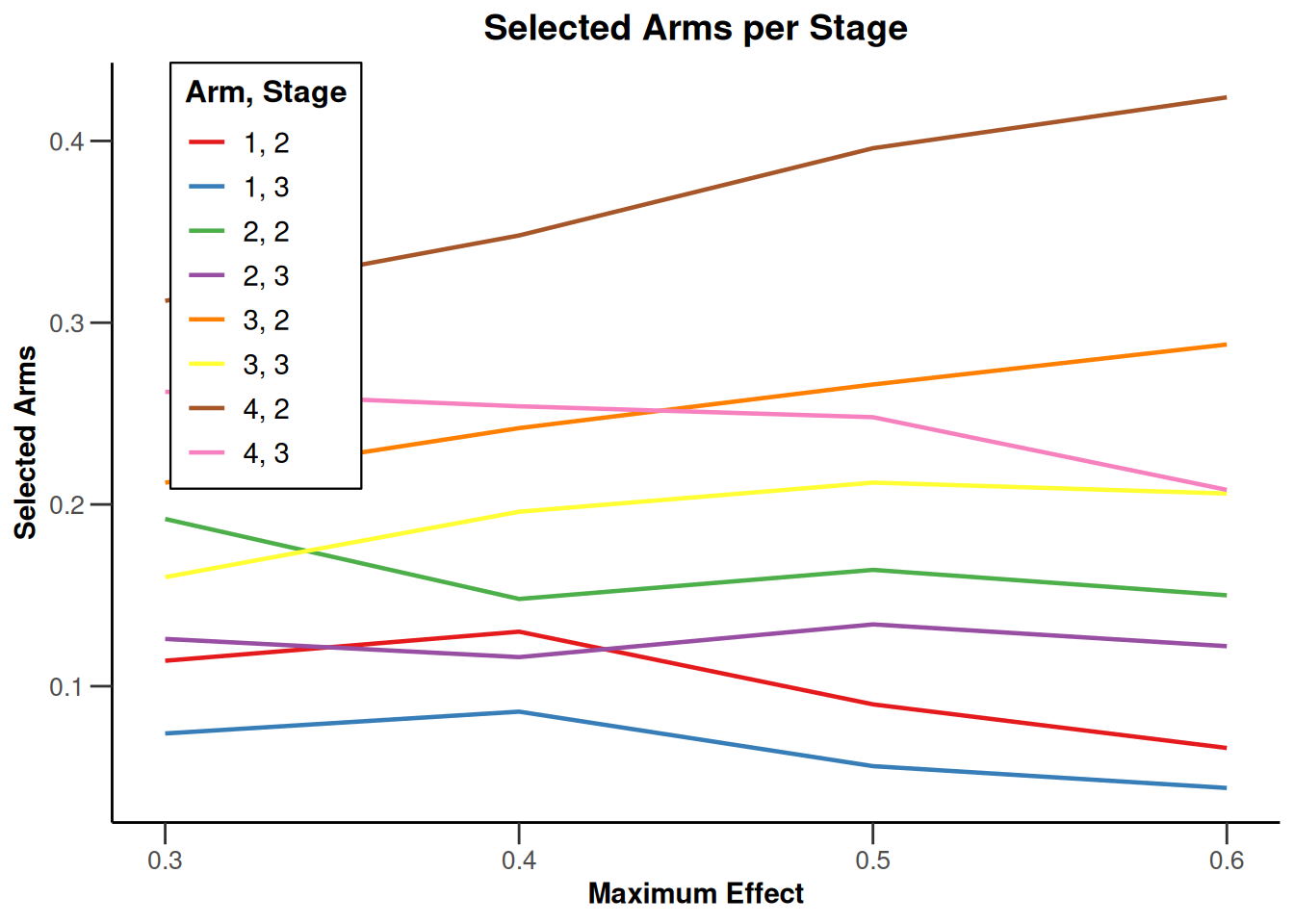
Simulation results multi-arm
Simulation results multi-arm - means
getDesignInverseNormal(
informationRates = c(0.2, 0.6, 1),
futilityBounds = c(-0.5, 0.5)
) |>
getSimulationMultiArmMeans(
typeOfShape = "linear",
activeArms = 4,
plannedSubjects = c(10, 30, 50),
stDev = 1.2,
muMaxVector = seq(0.3, 0.6, 0.1),
adaptations = rep(TRUE, 2),
conditionalPower = 0.8,
minNumberOfSubjectsPerStage = c(10, 4, 4),
maxNumberOfSubjectsPerStage = c(10, 100, 100),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)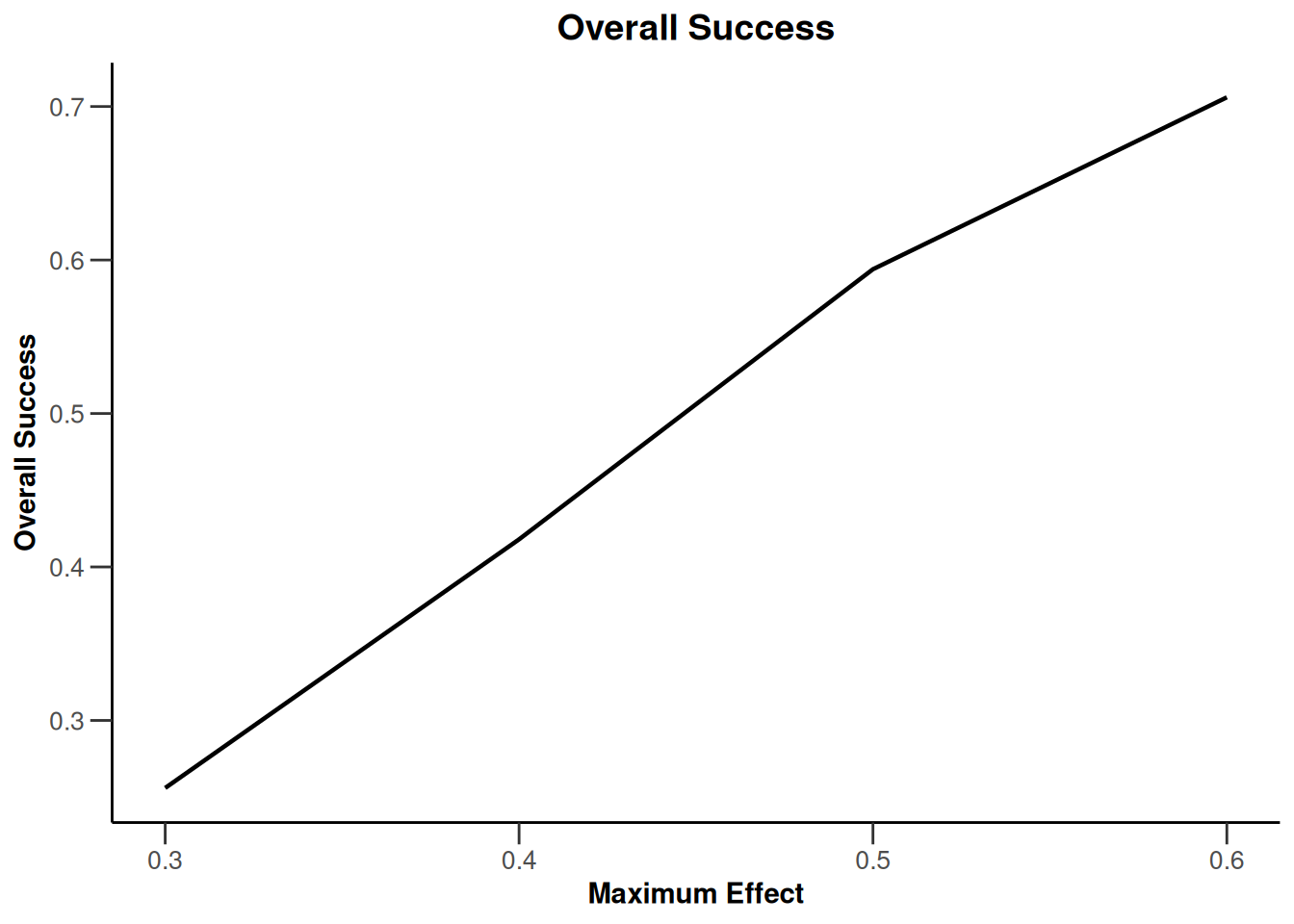

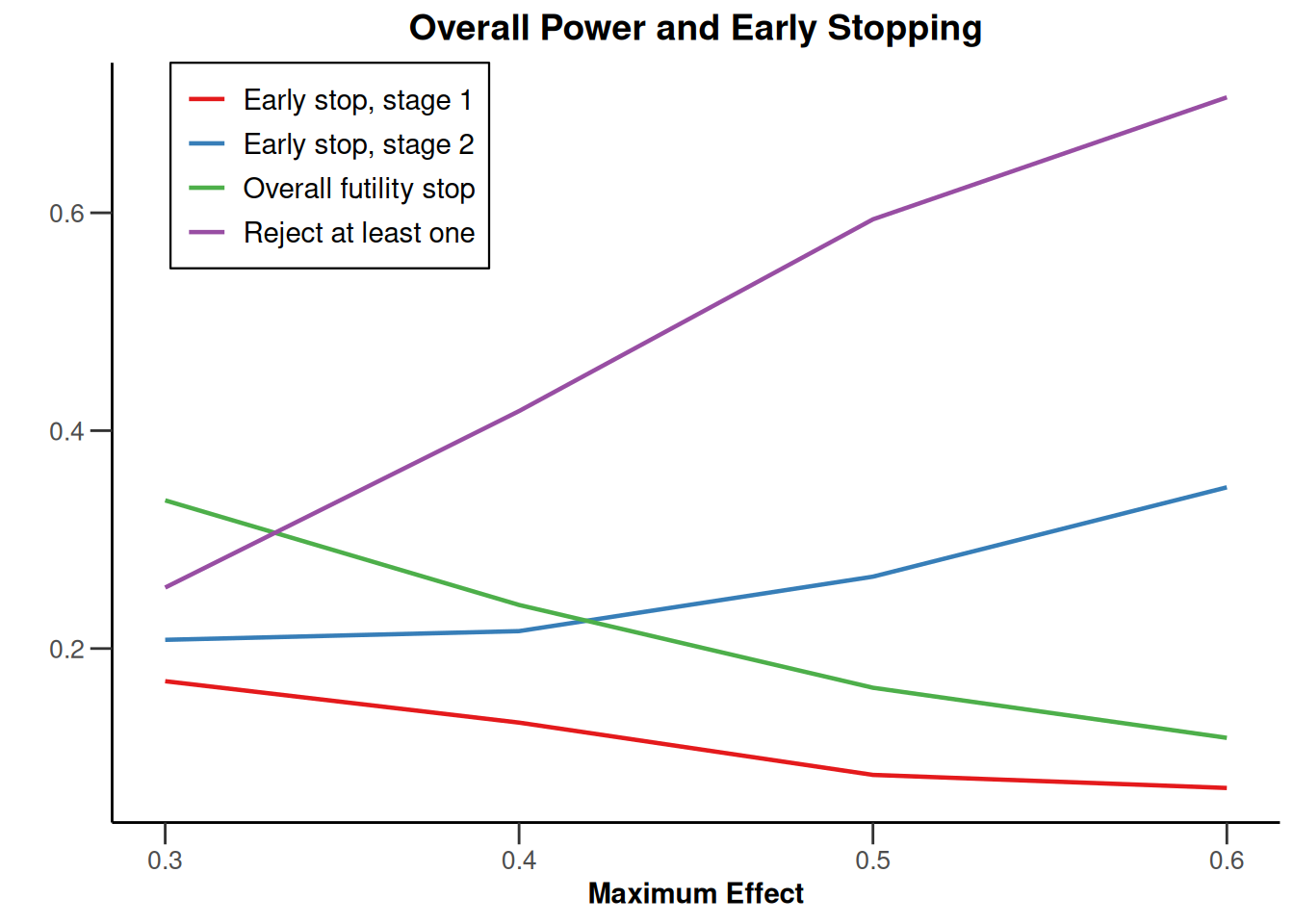
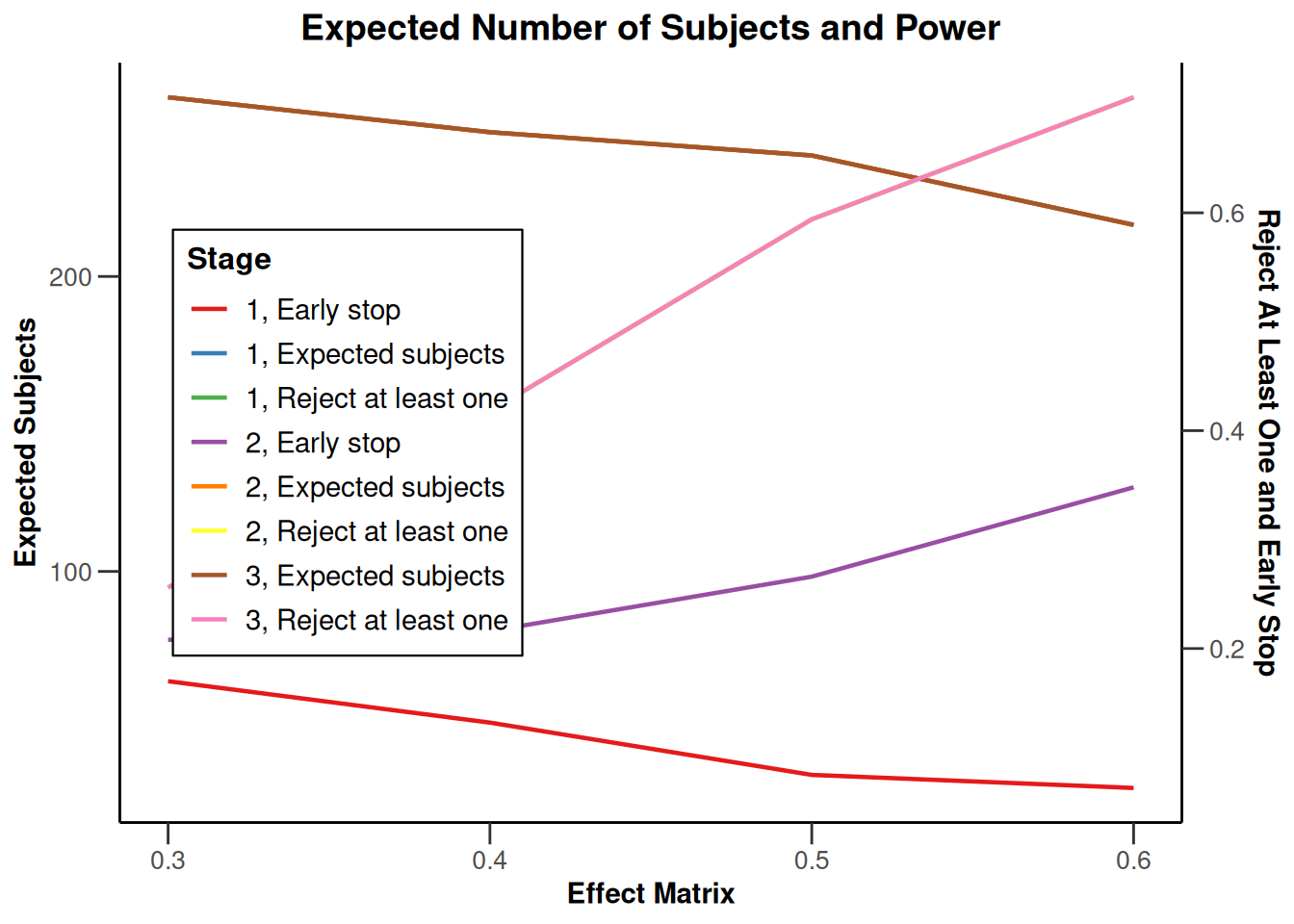
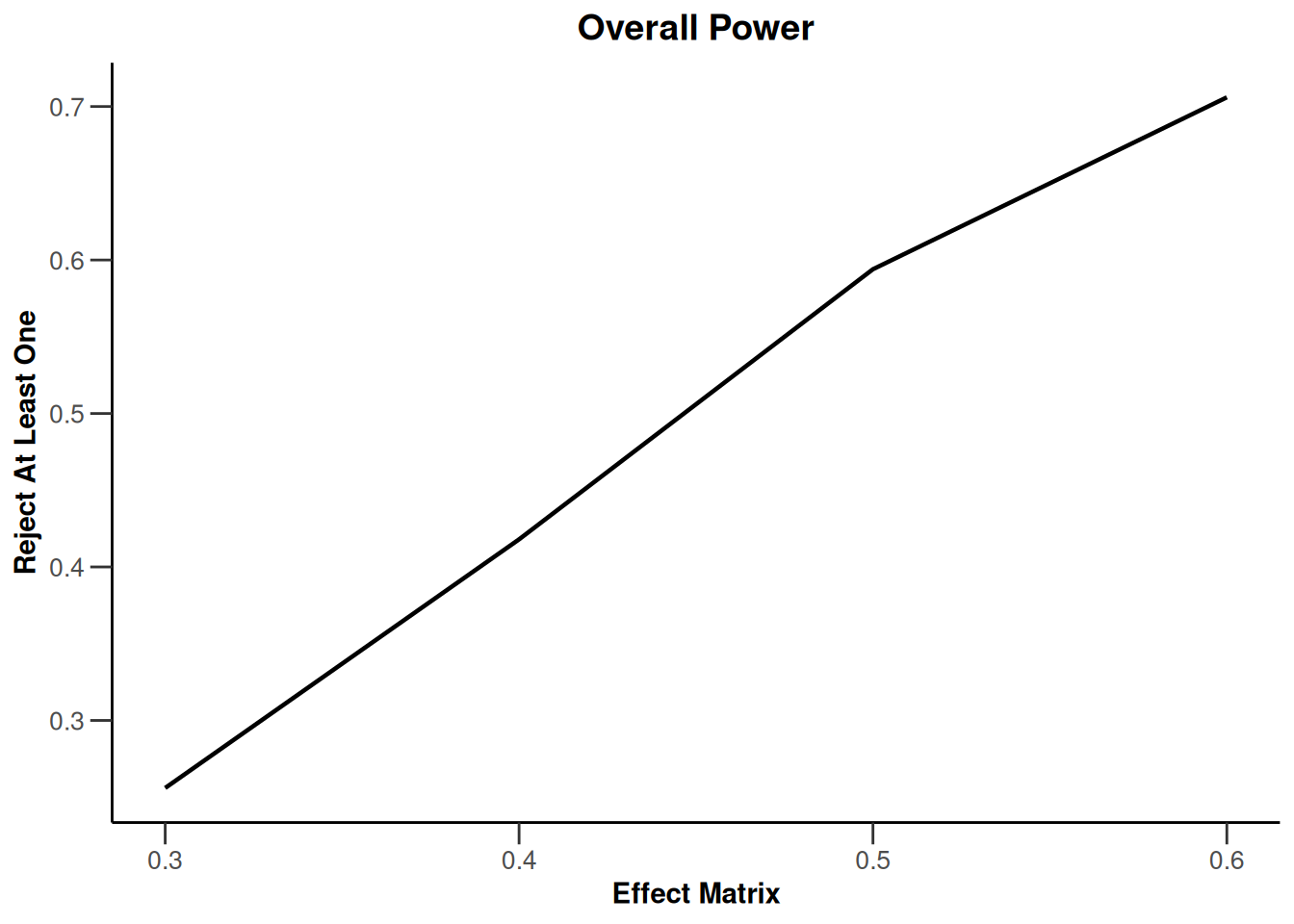
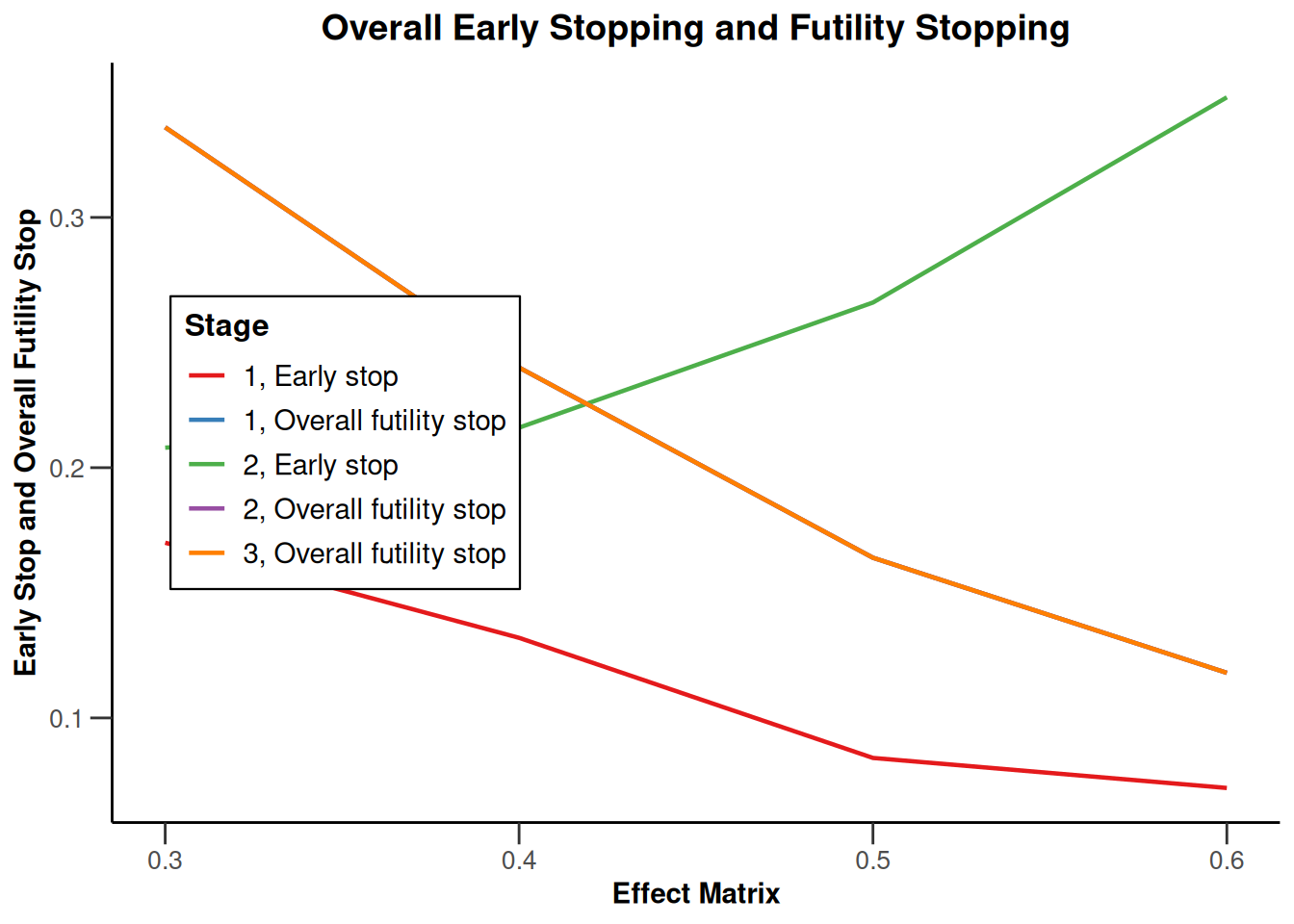
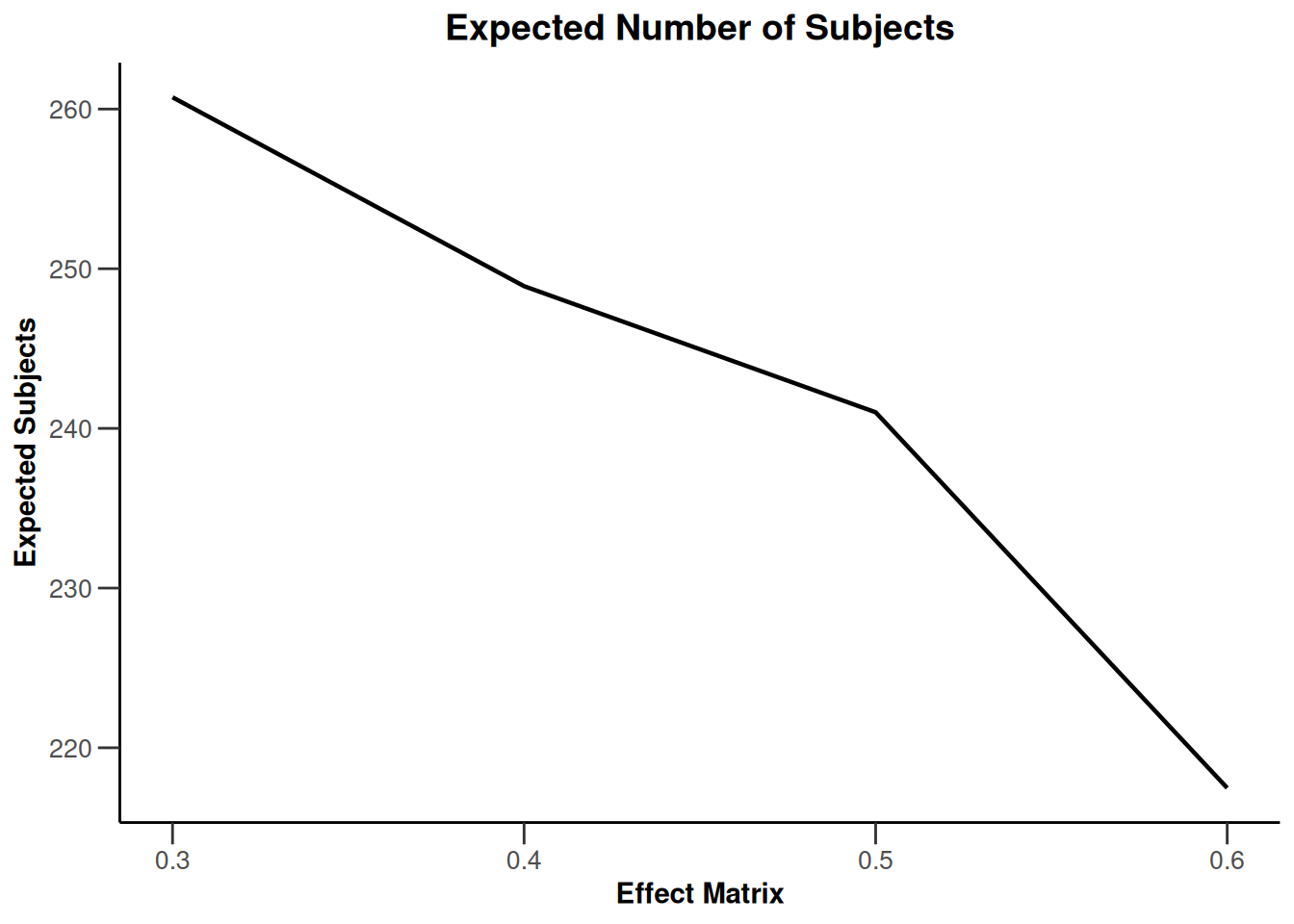
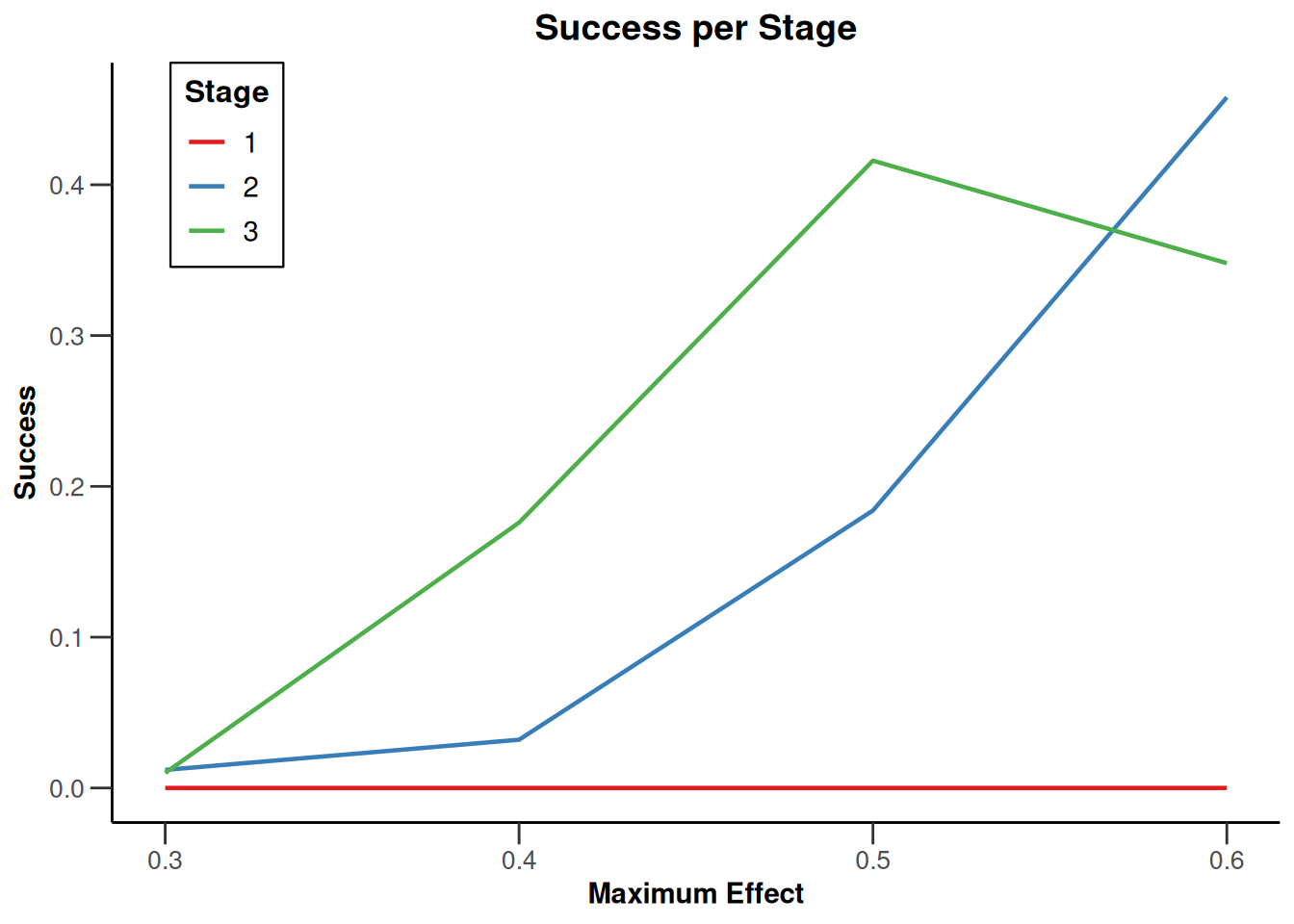
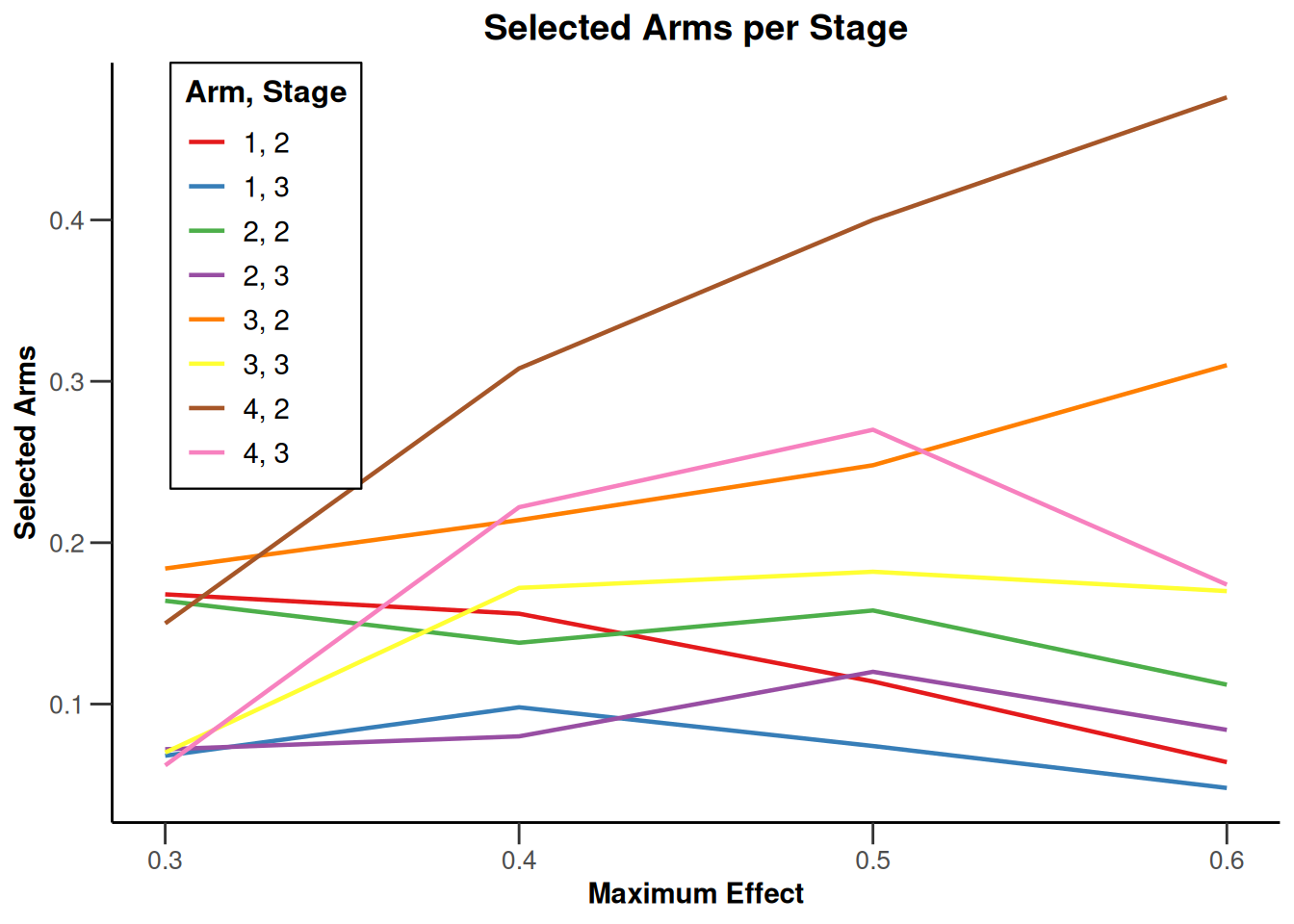
Simulation results multi-arm - rates
getDesignInverseNormal(
informationRates = c(0.2, 0.6, 1),
futilityBounds = c(-0.5, 0.5)
) |>
getSimulationMultiArmRates(
typeOfShape = "linear",
activeArms = 4,
plannedSubjects = c(10, 30, 50),
piControl = 0.3,
piMaxVector = seq(0.3, 0.6, 0.1),
adaptations = rep(TRUE, 2),
conditionalPower = 0.8,
minNumberOfSubjectsPerStage = c(10, 4, 4),
maxNumberOfSubjectsPerStage = c(10, 100, 100),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)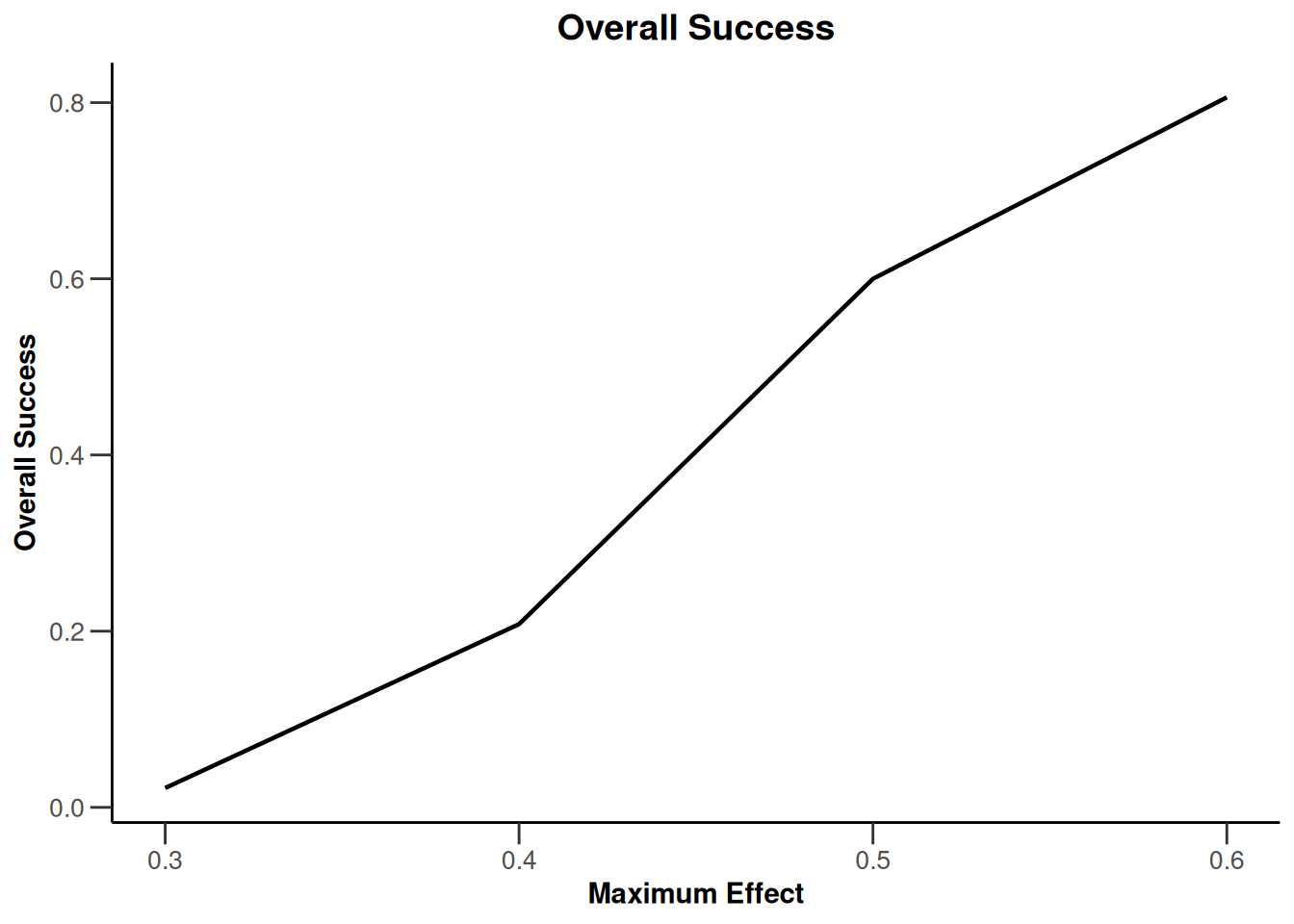
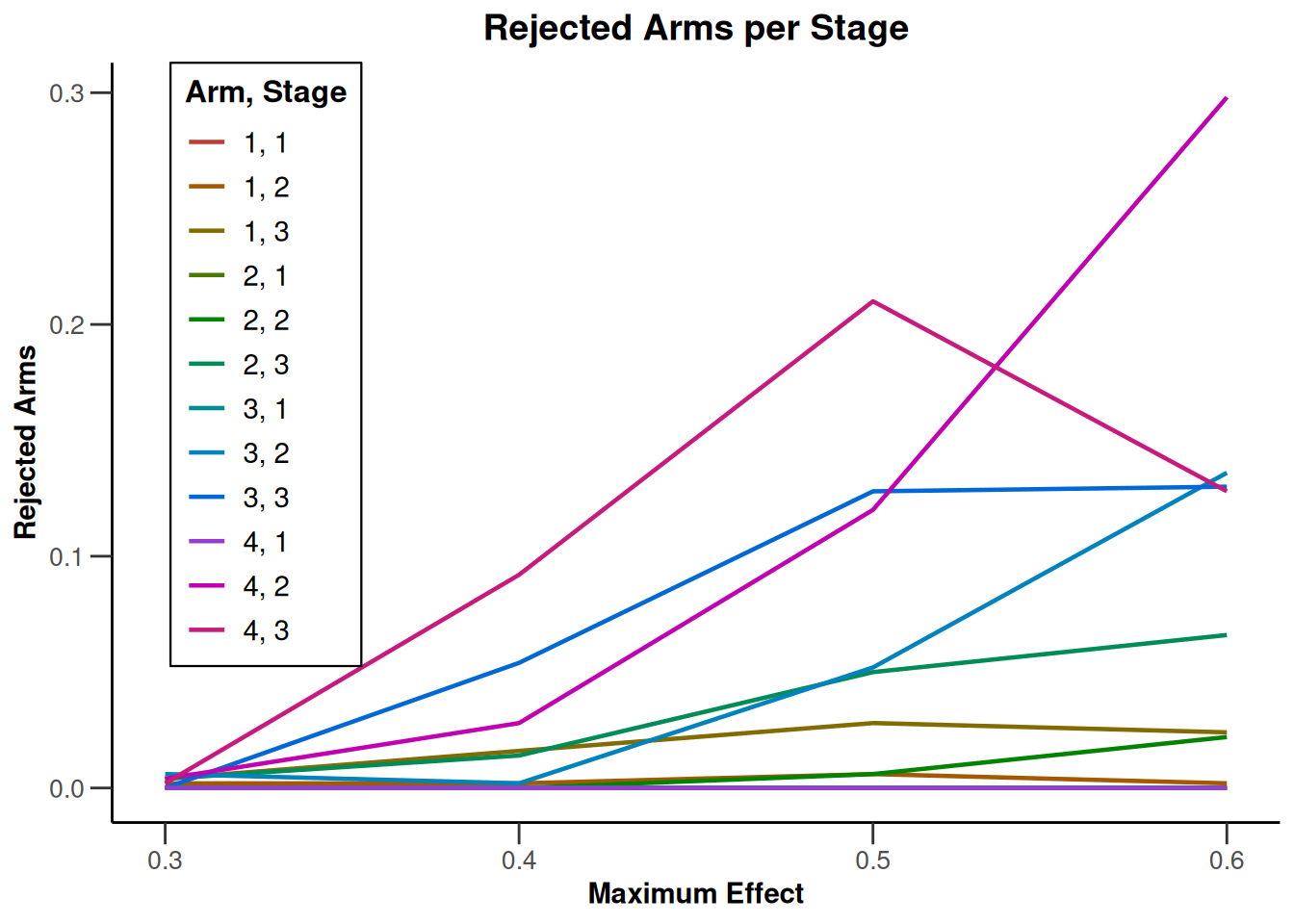
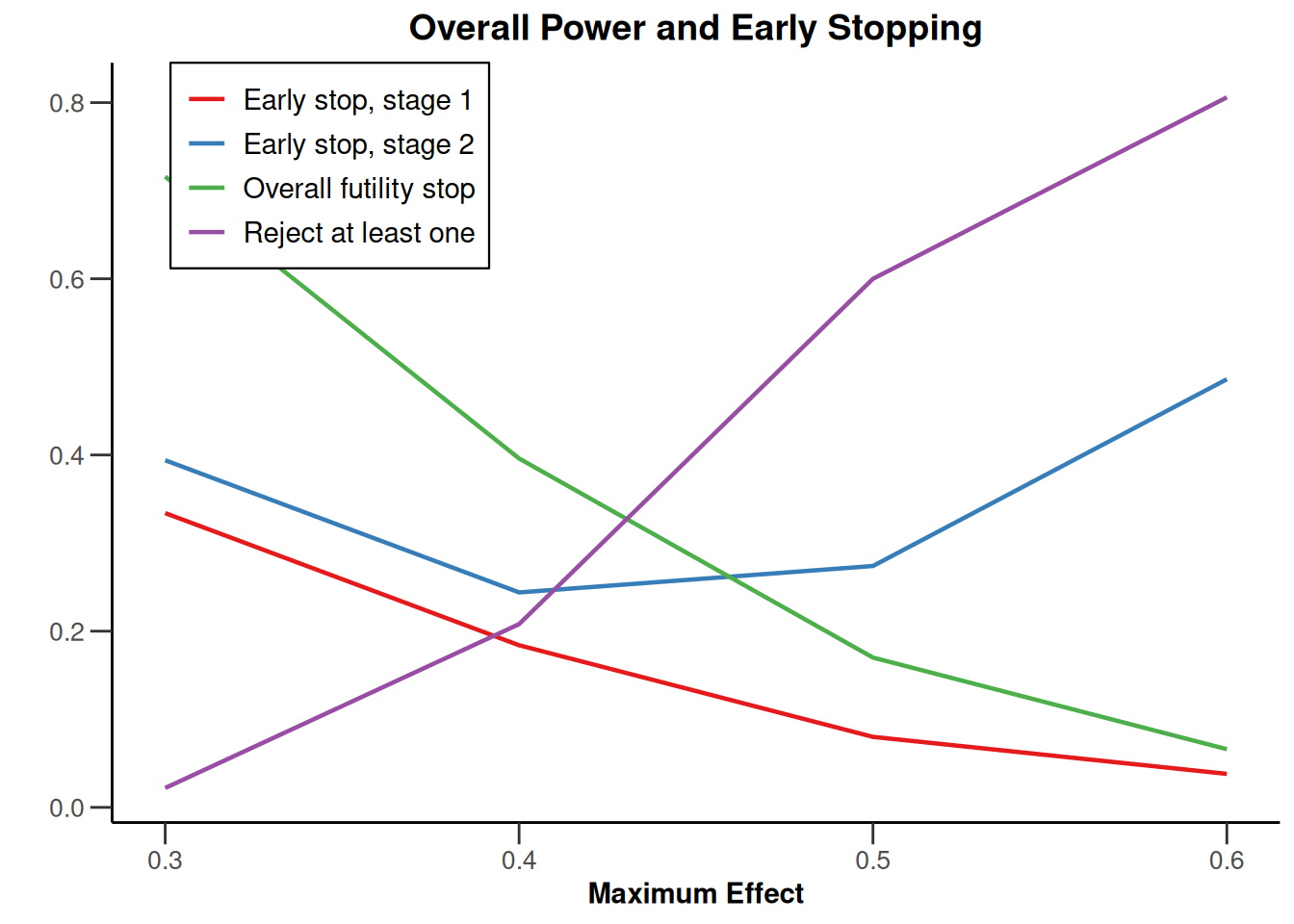
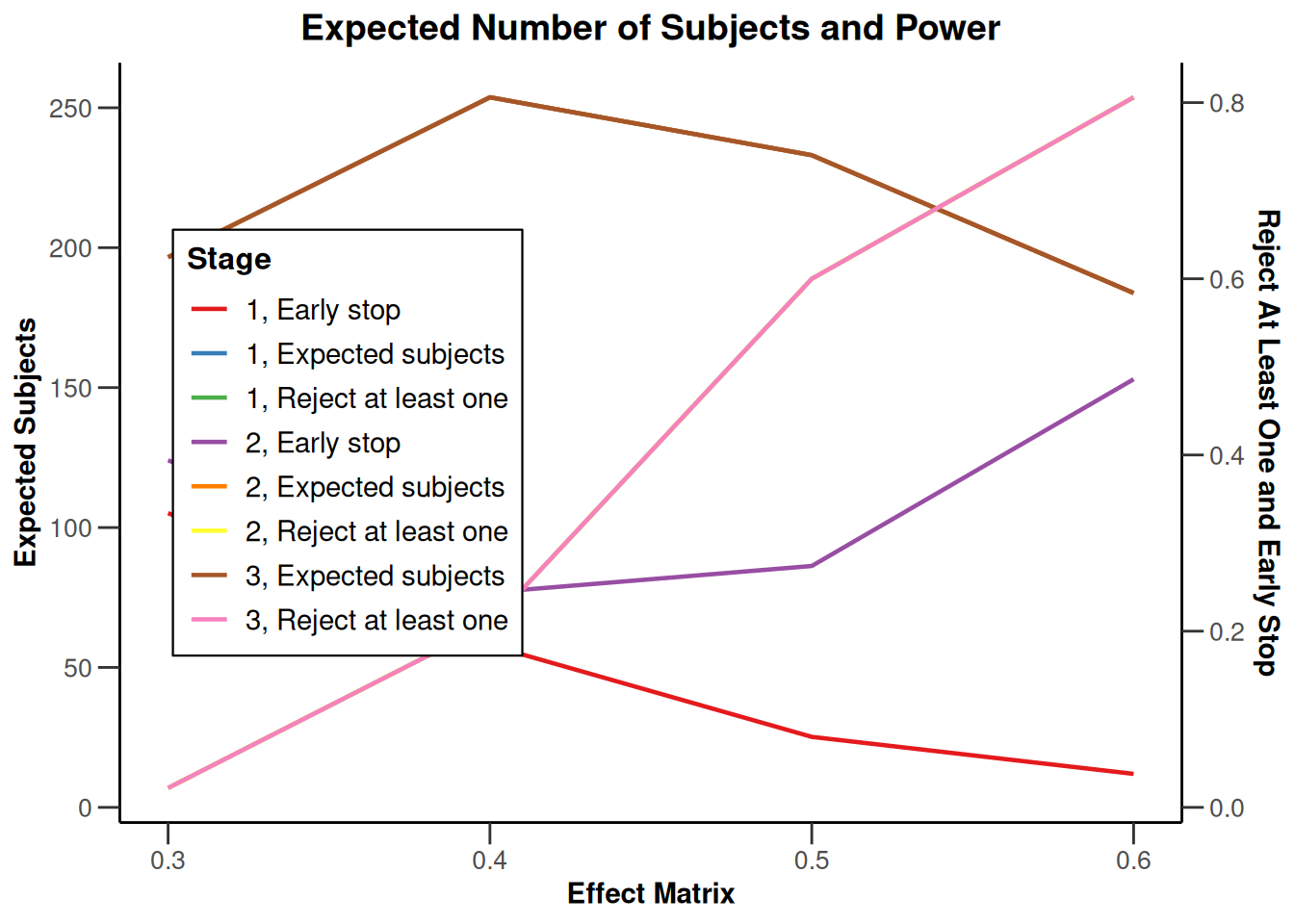
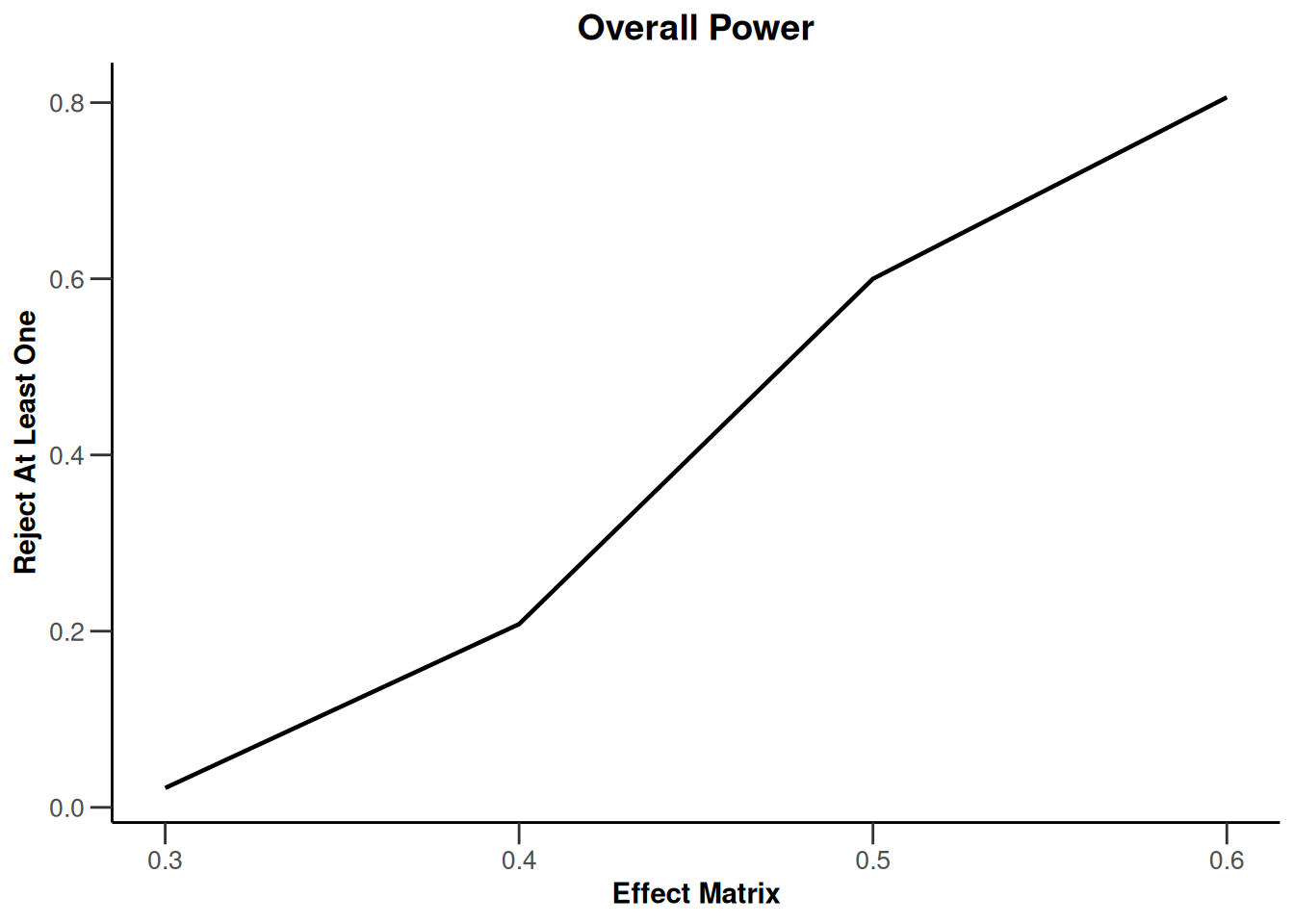
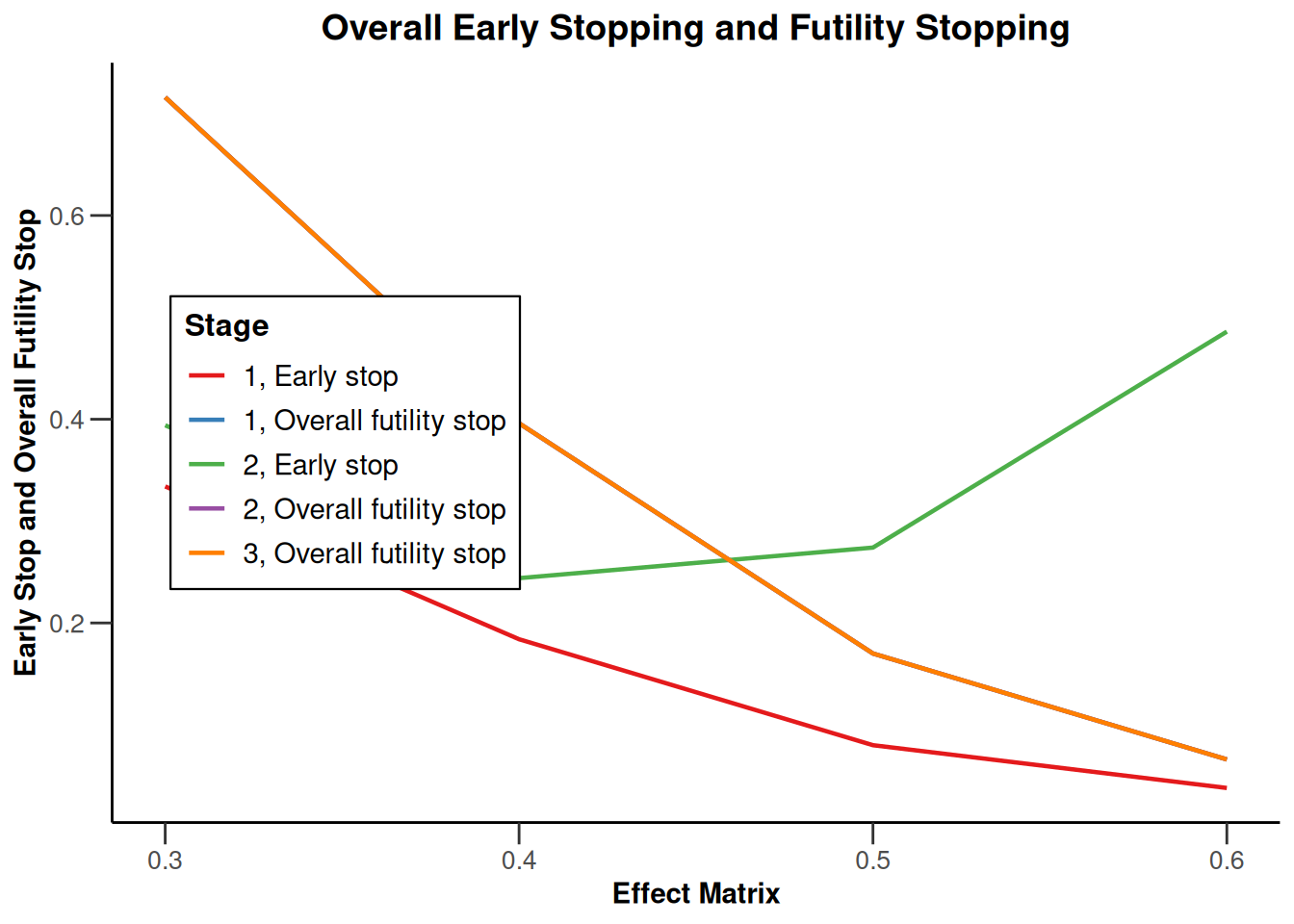
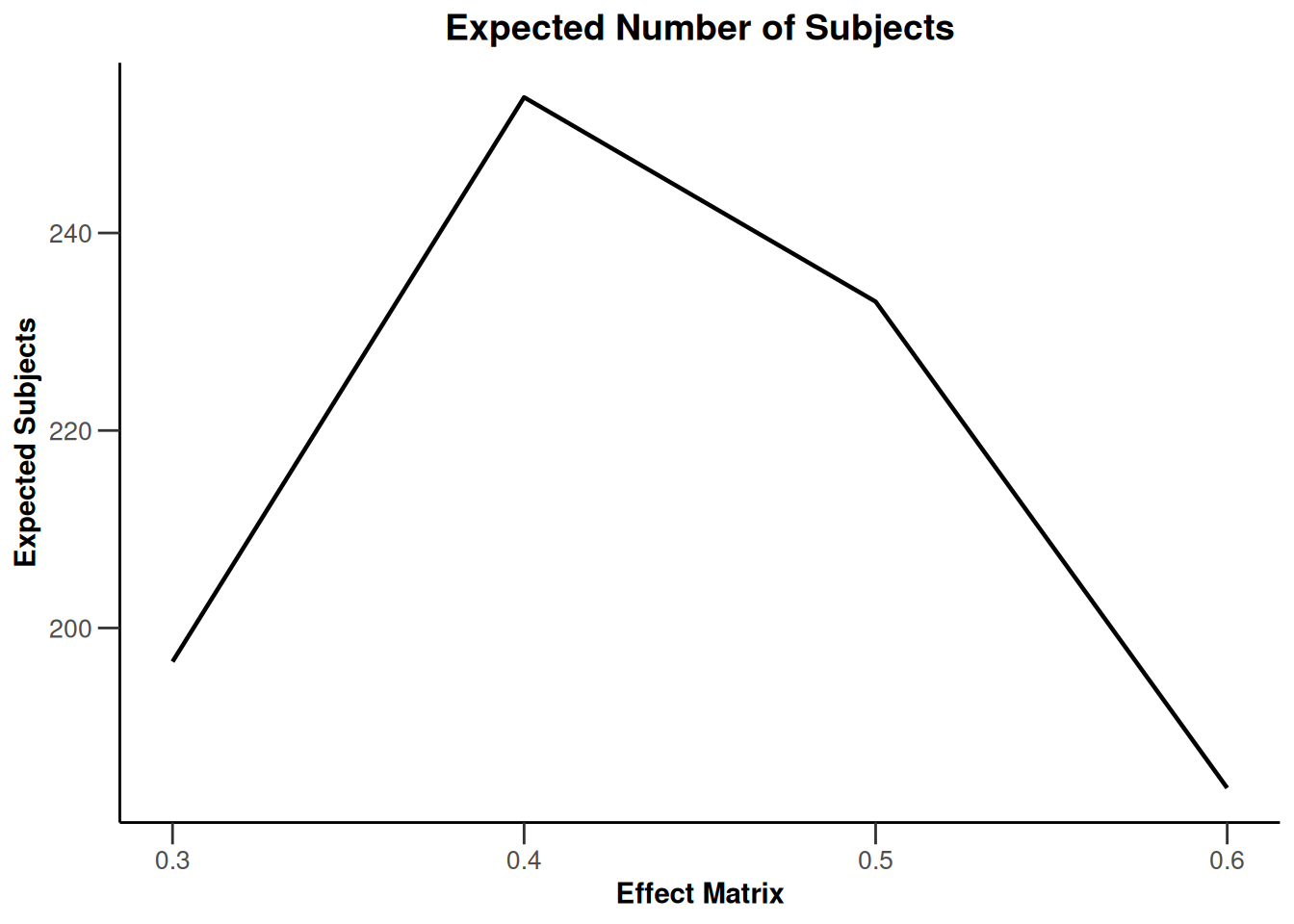
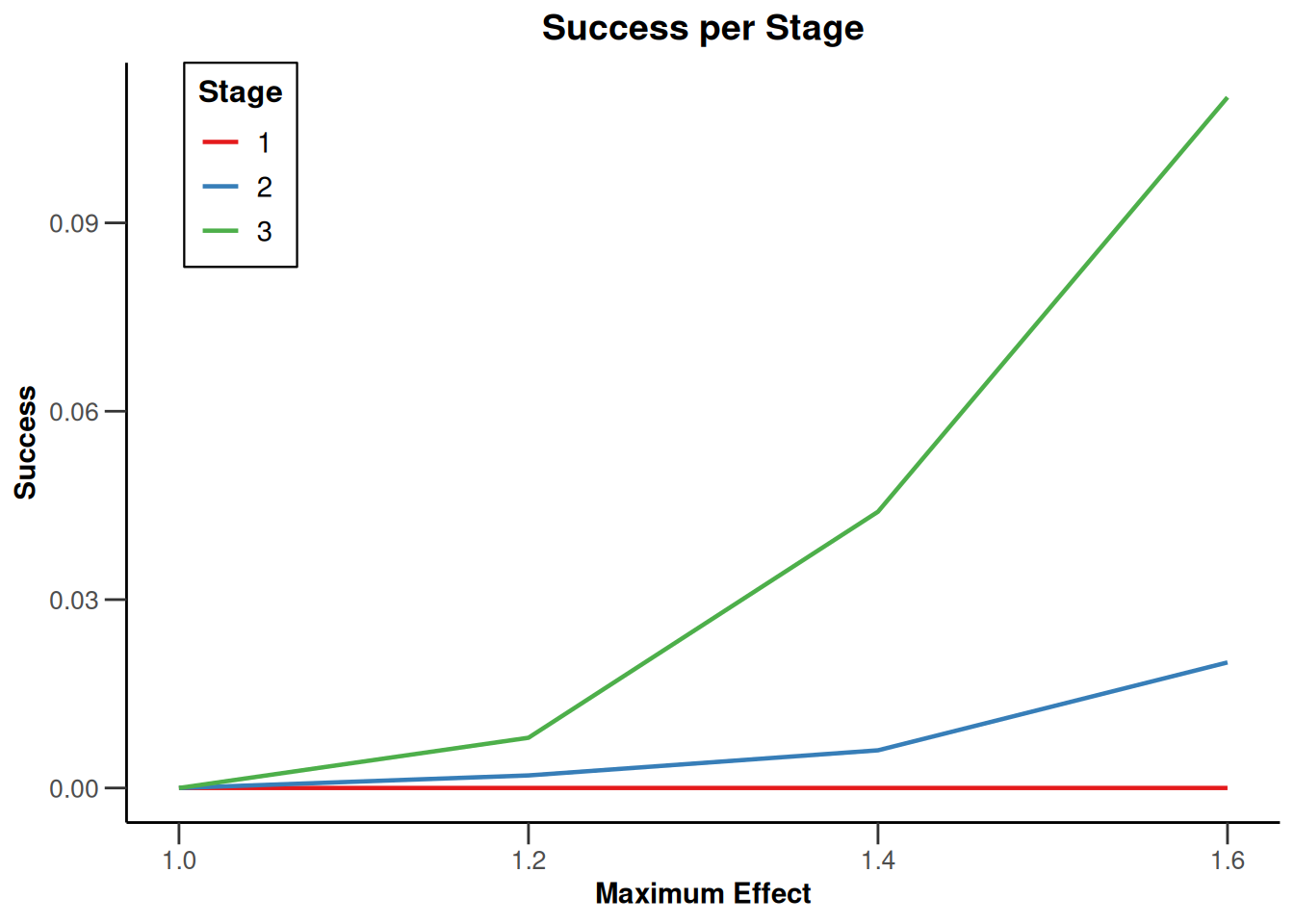
Simulation results multi-arm - survival
getDesignInverseNormal(
informationRates = c(0.2, 0.6, 1),
futilityBounds = c(-0.5, 0.5)
) |>
getSimulationMultiArmSurvival(
activeArms = 4,
typeOfSelection = "rBest",
rValue = 2,
plannedEvents = c(10, 30, 50),
omegaMaxVector = seq(1, 1.6, 0.2),
adaptations = rep(TRUE, 2),
conditionalPower = 0.8,
minNumberOfEventsPerStage = c(10, 4, 4),
maxNumberOfEventsPerStage = c(10, 100, 100),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)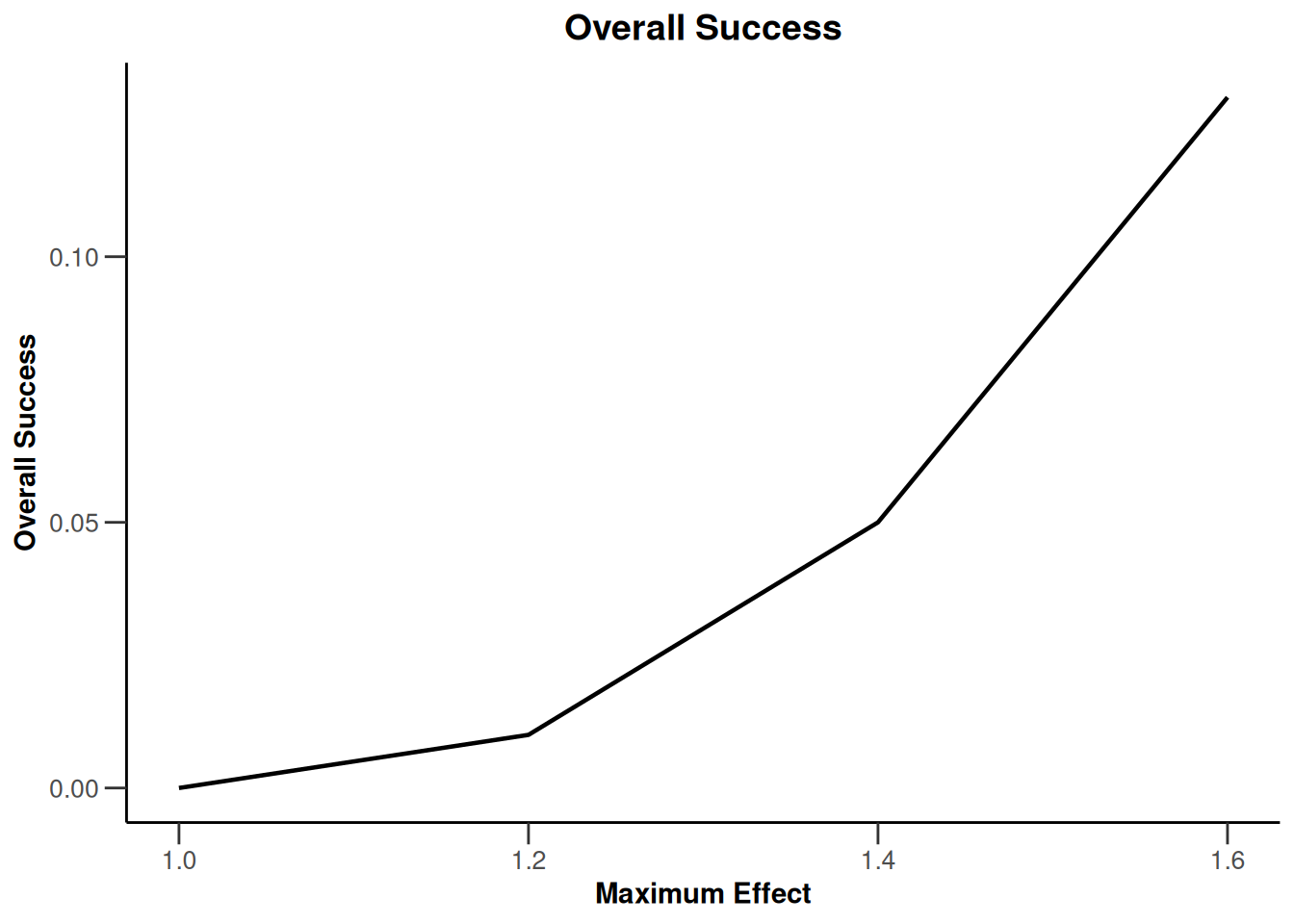
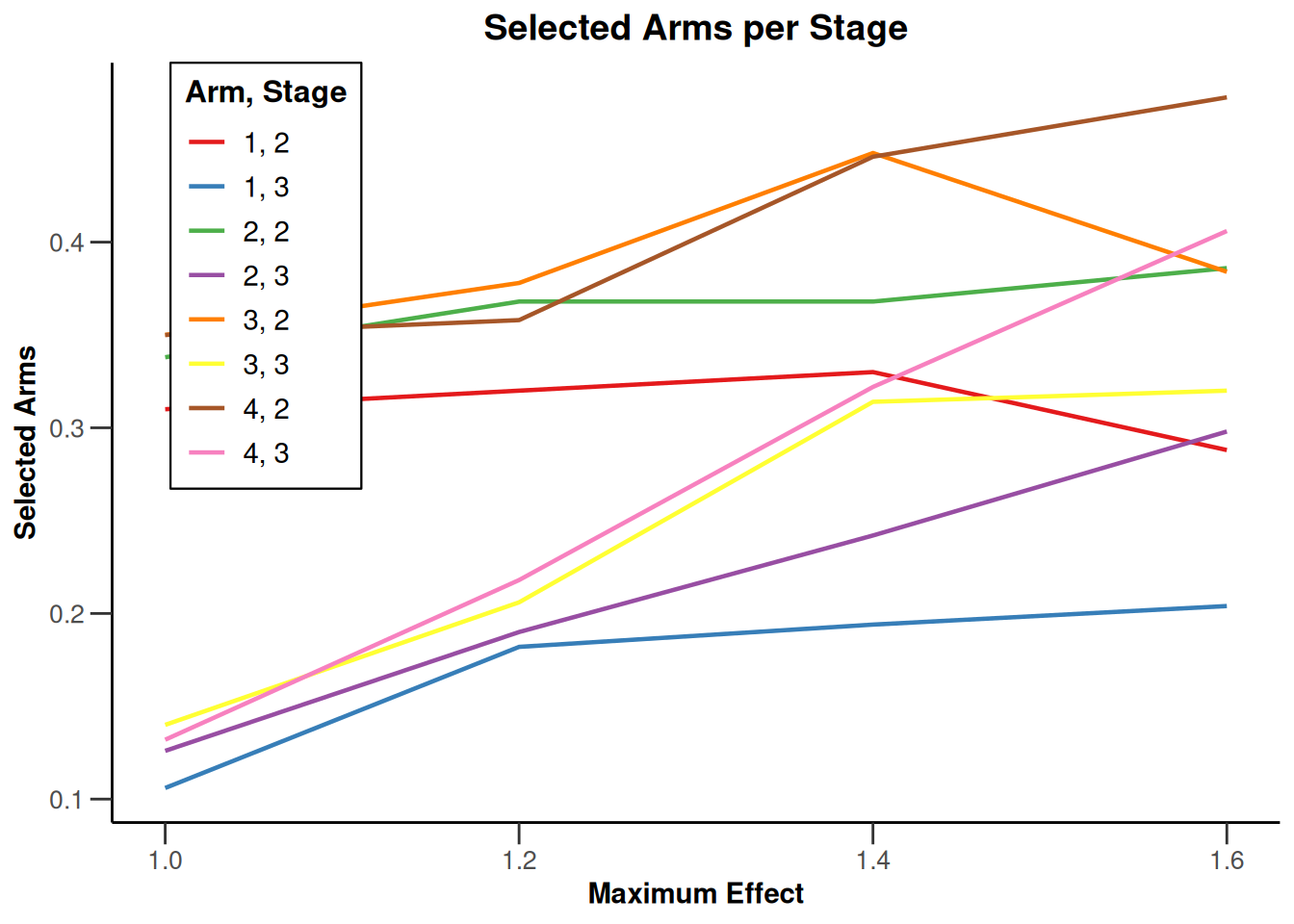
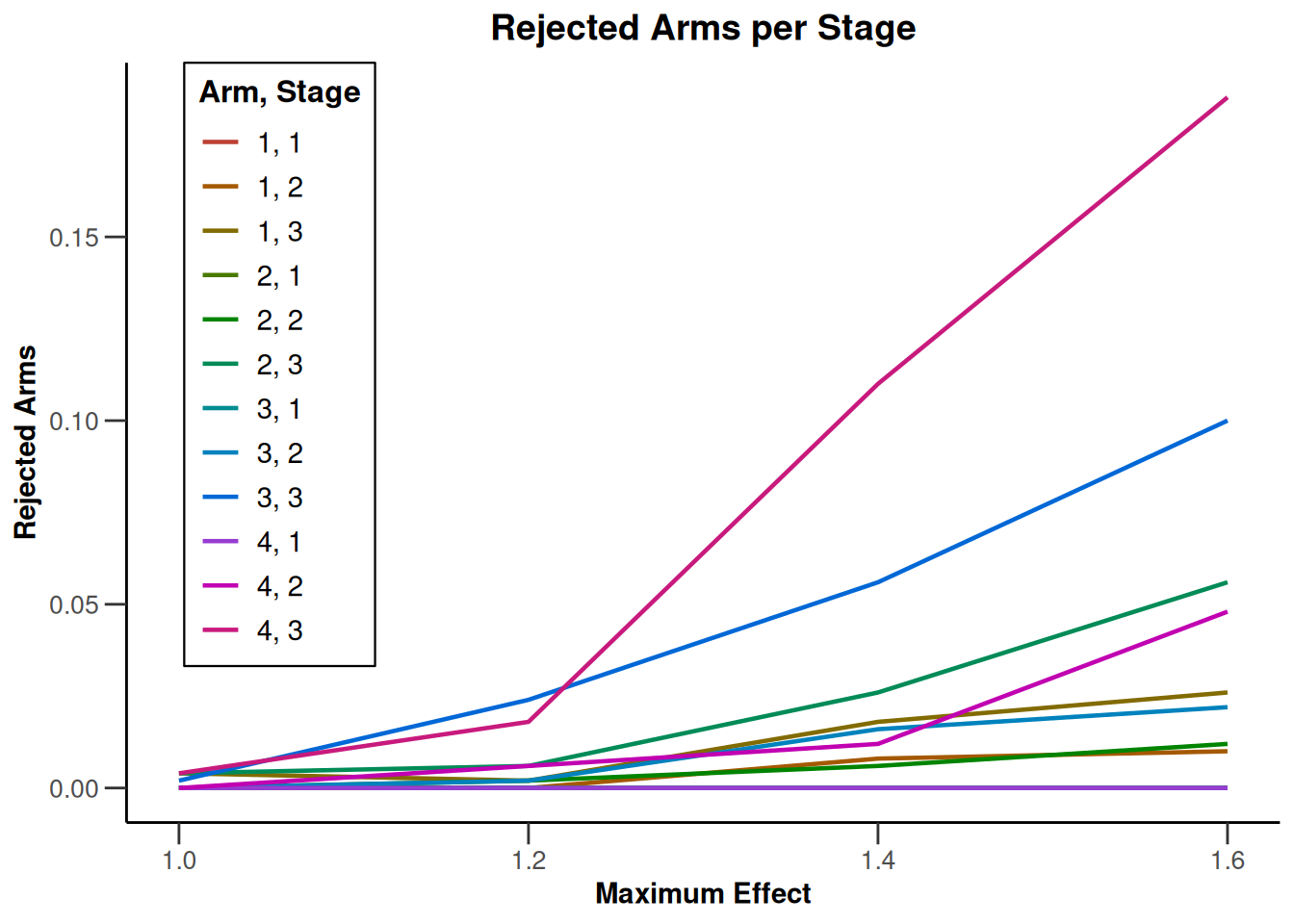
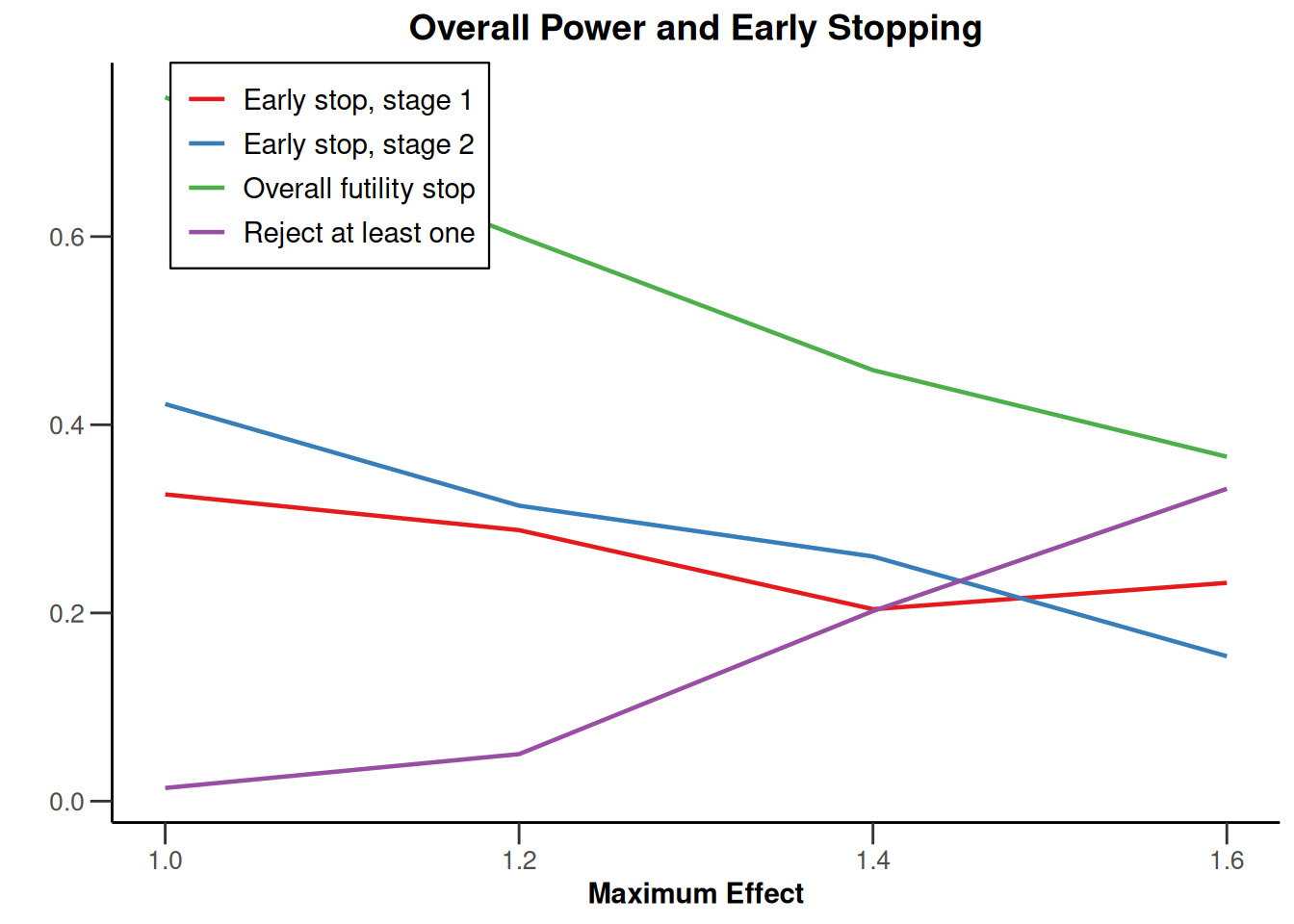
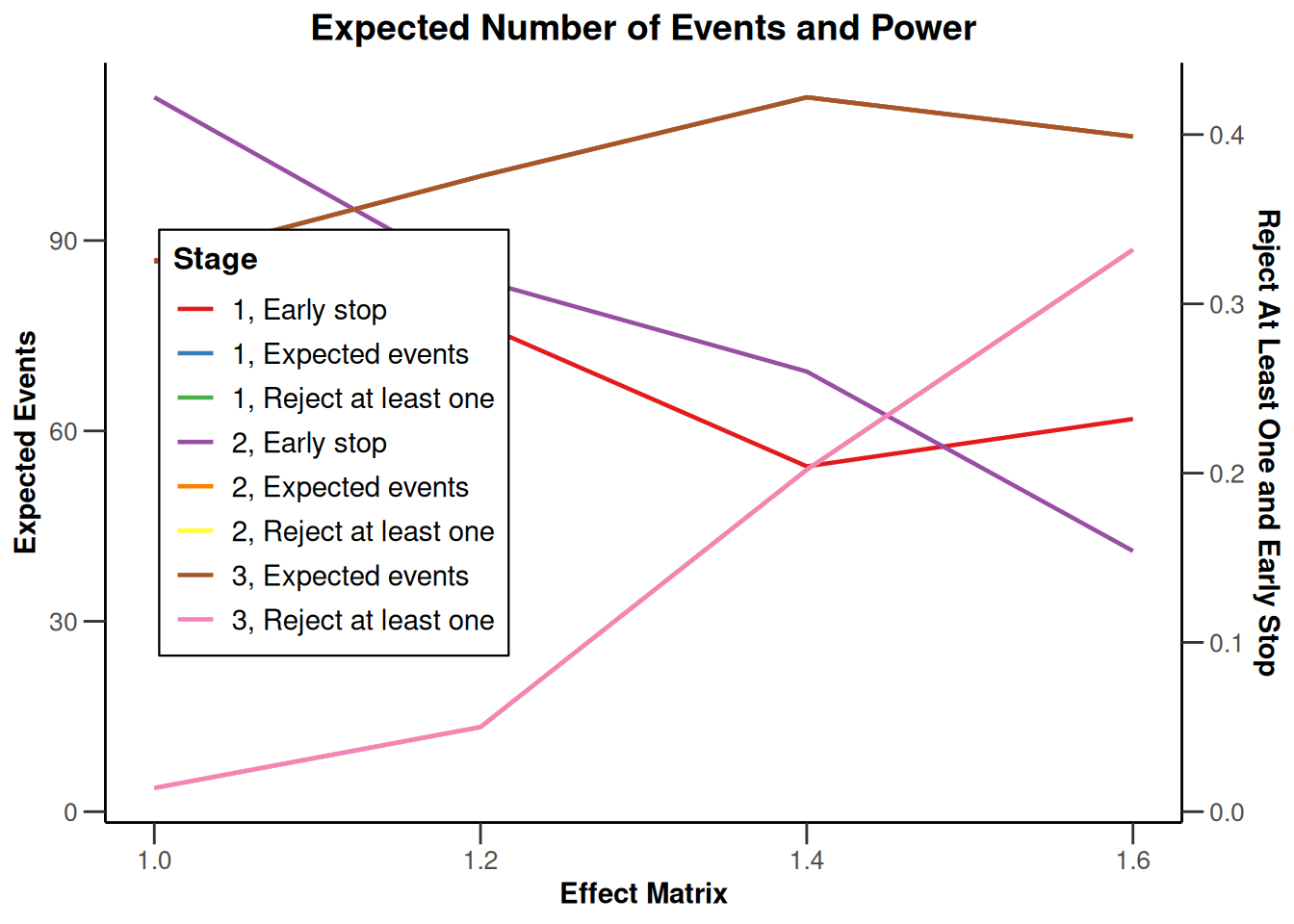
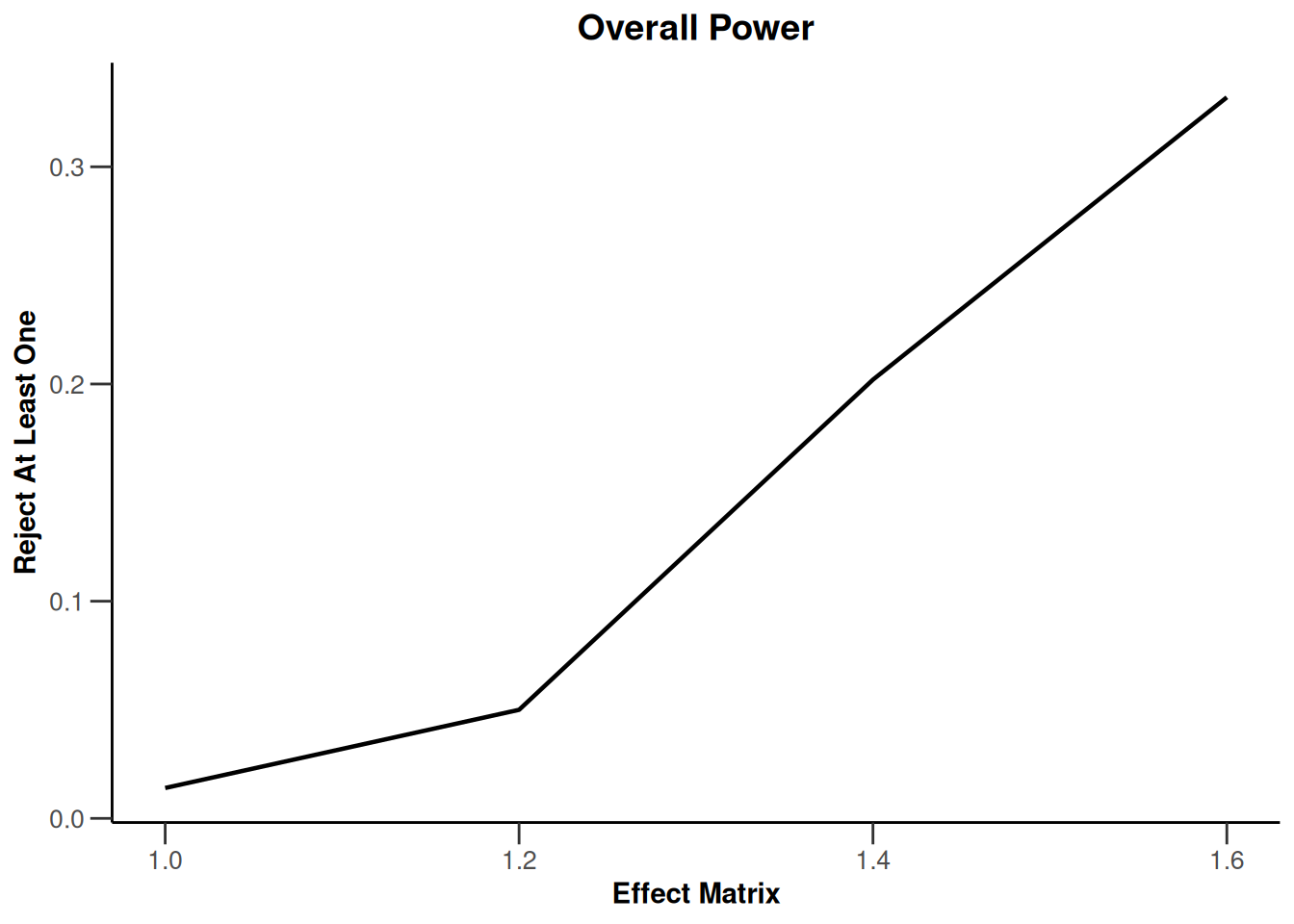
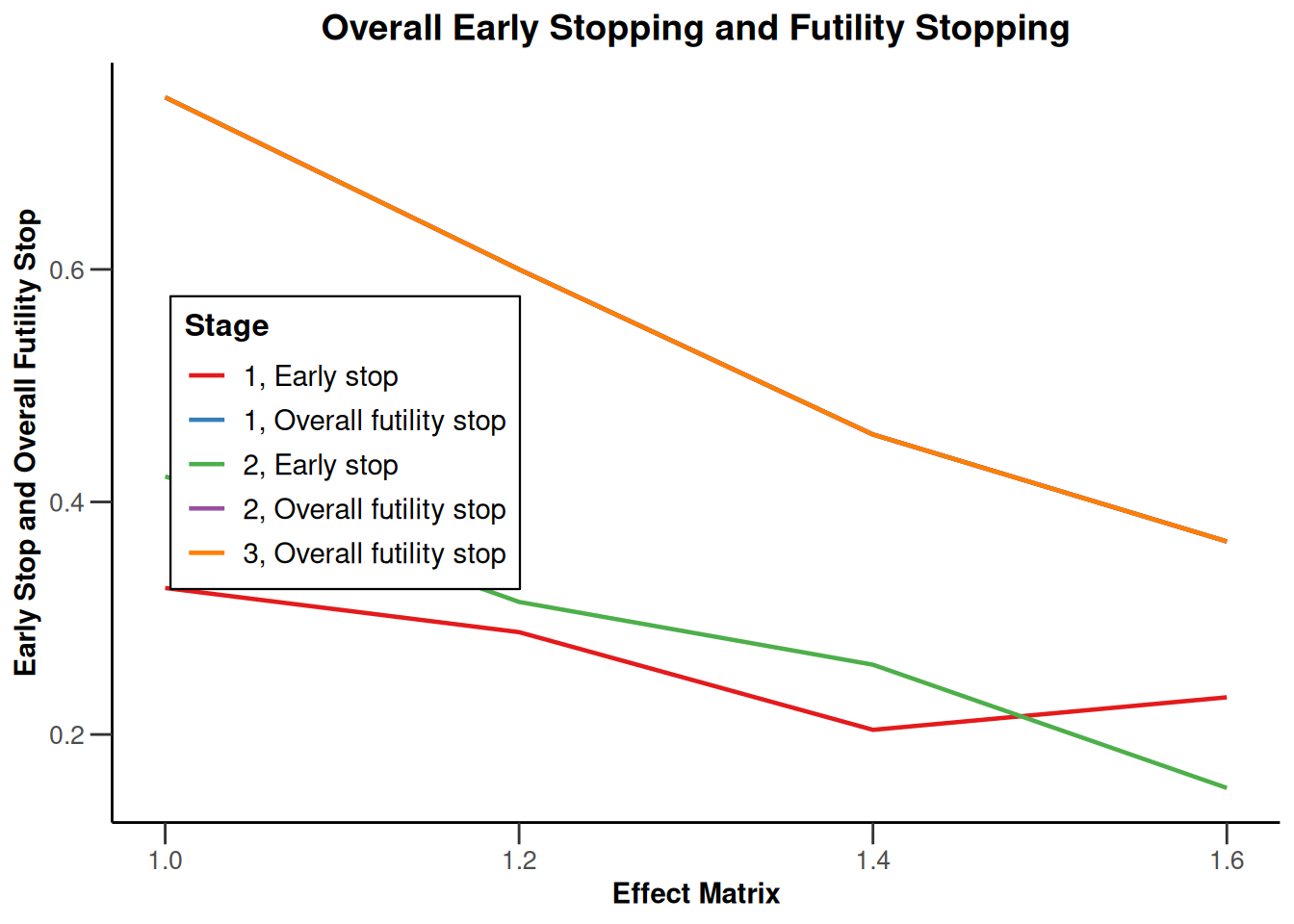
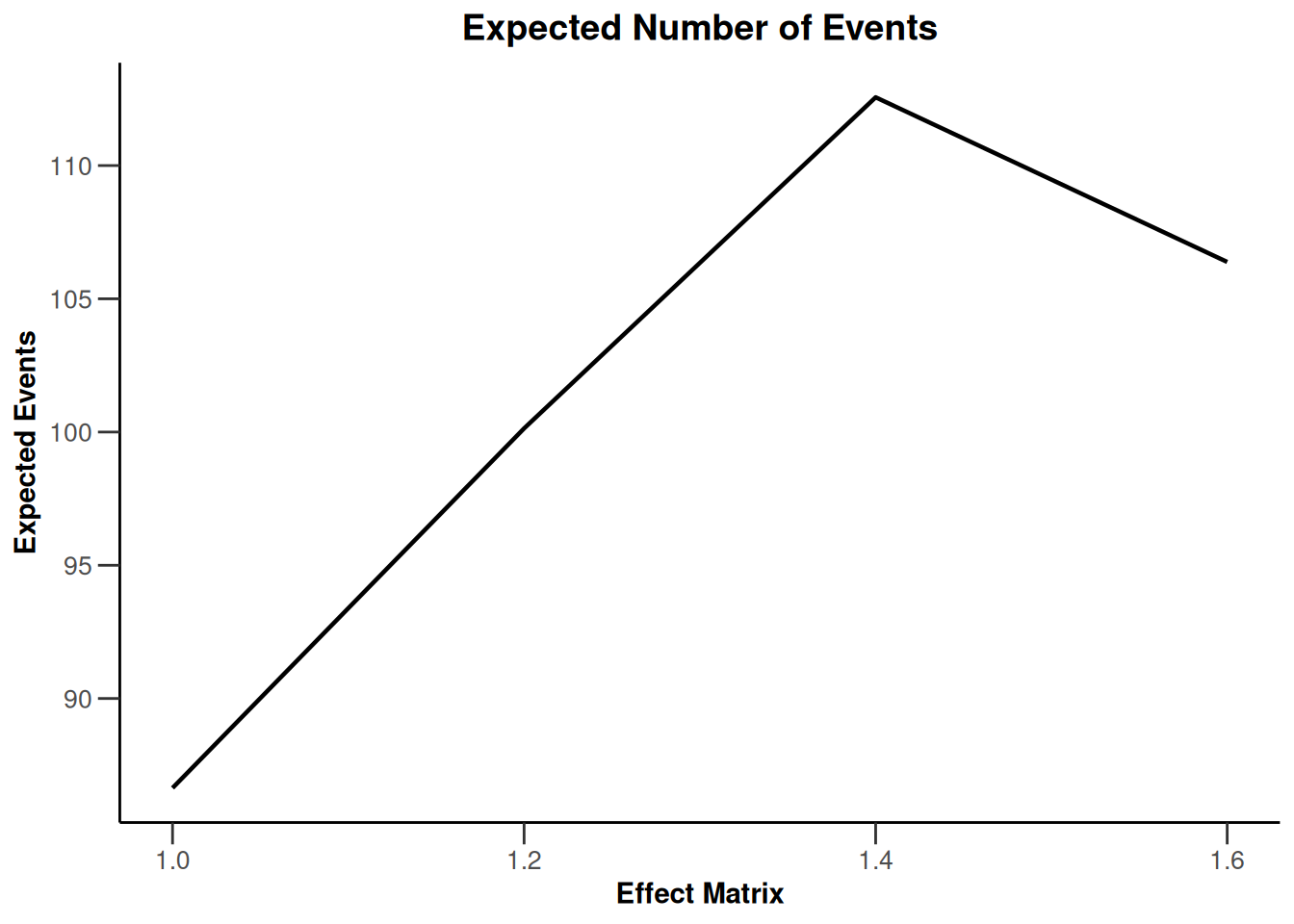
Simulation results enrichment
Simulation results enrichment - means
design <- getDesignInverseNormal(
informationRates = c(0.2, 0.6, 1),
futilityBounds = c(-0.5, 0.5)
)
# Define subgroups and their prevalences
subGroups <- c("S1", "S12", "S2", "R") # fixed names!
prevalences <- c(0.2, 0.3, 0.4, 0.1)
effectR <- 1.5
effectS12 <- 5
m <- c()
for (effectS1 in seq(0, 5, 5)) {
for (effectS2 in seq(0, 5, 5)) {
m <- c(m, effectS1, effectS12, effectS2, effectR)
}
}
effects <- matrix(m, byrow = TRUE, ncol = 4)
stDev <- 10
# Define effect list
el <- list(
subGroups = subGroups, prevalences = prevalences,
stDevs = stDev, effects = effects
)
# Perform simulation
design |>
getSimulationEnrichmentMeans(
plannedSubjects = c(10, 30, 50),
effectList = el,
adaptations = rep(TRUE, 2),
conditionalPower = 0.8,
minNumberOfSubjectsPerStage = c(10, 4, 4),
maxNumberOfSubjectsPerStage = c(10, 100, 100),
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)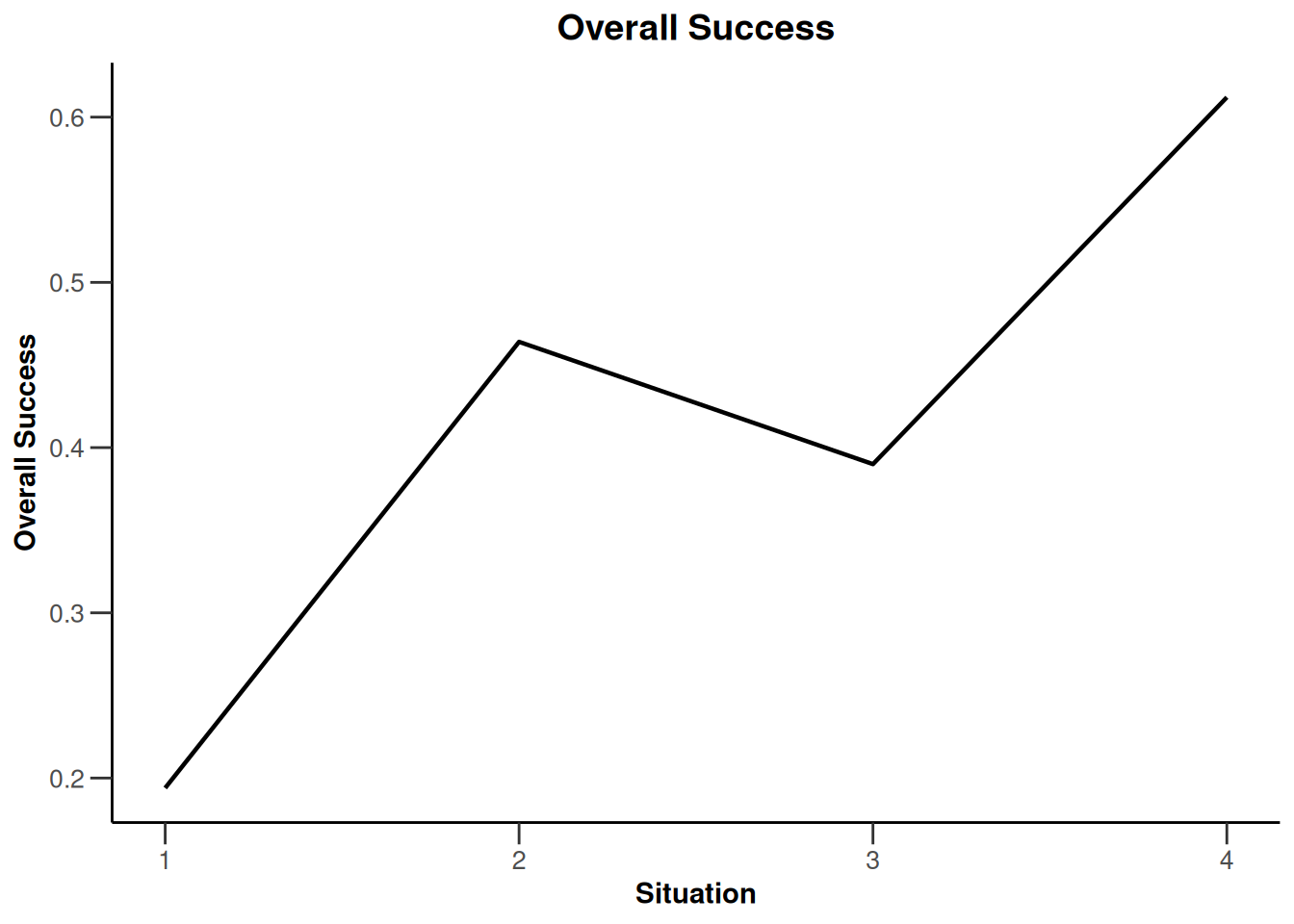
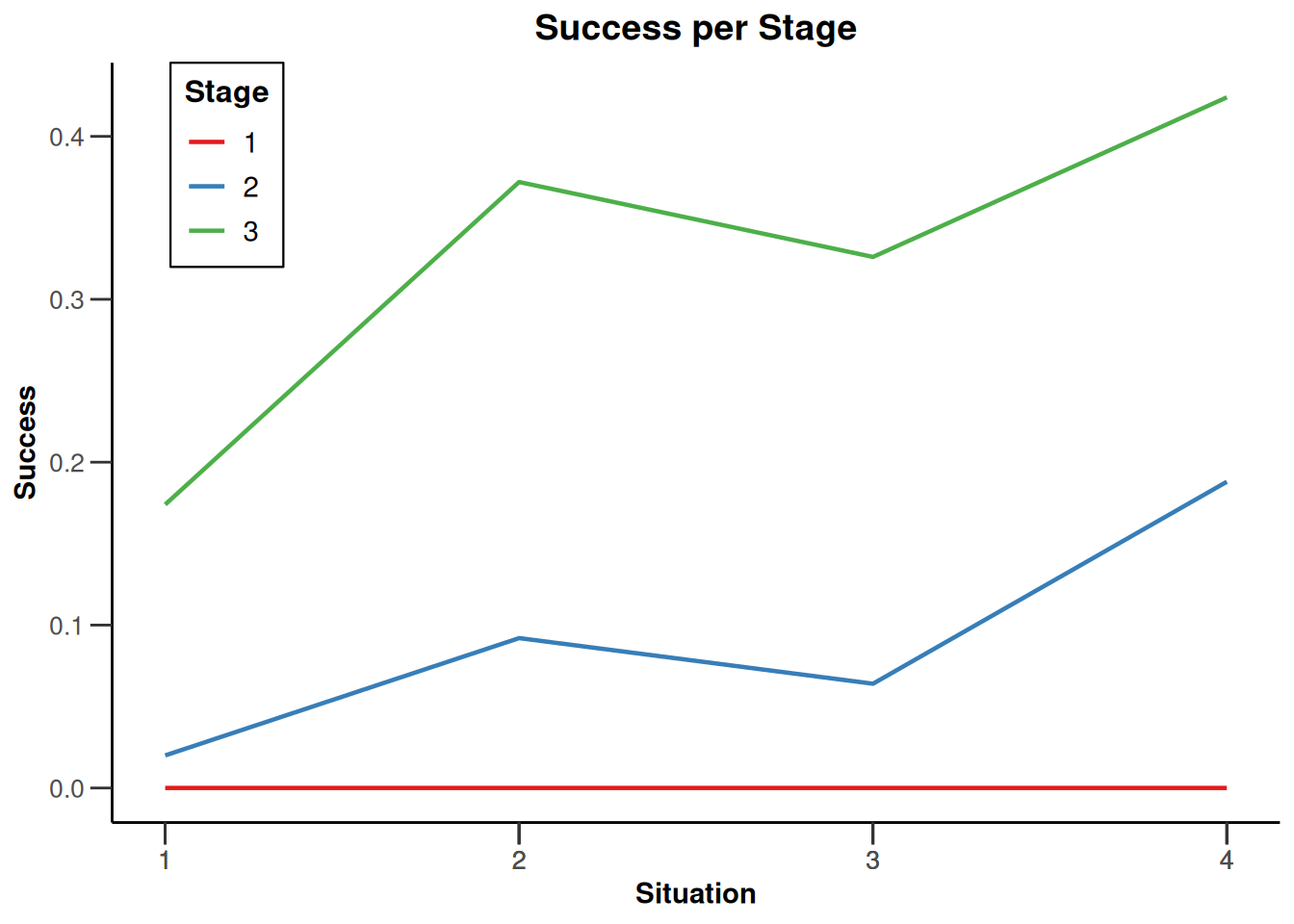
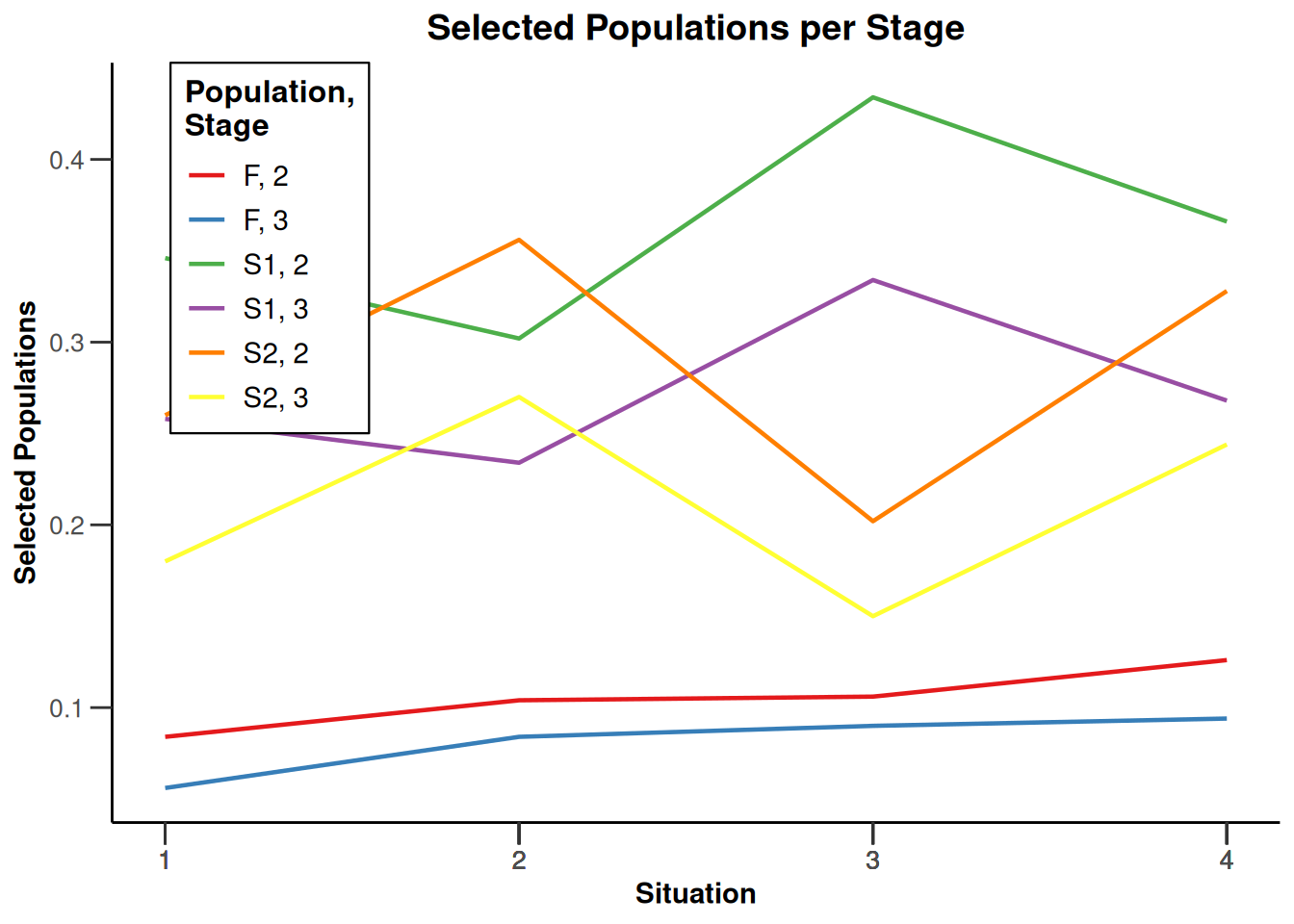
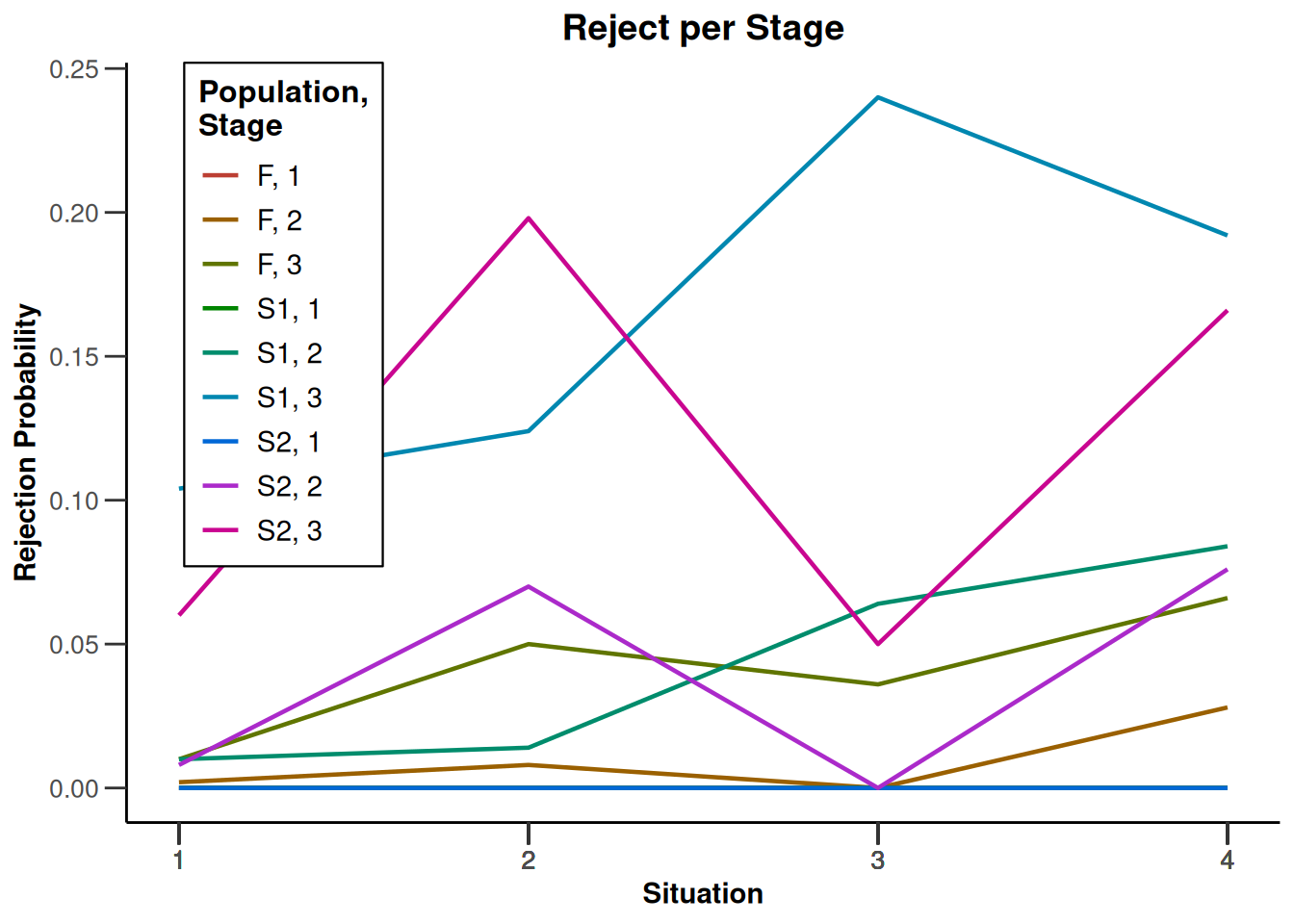
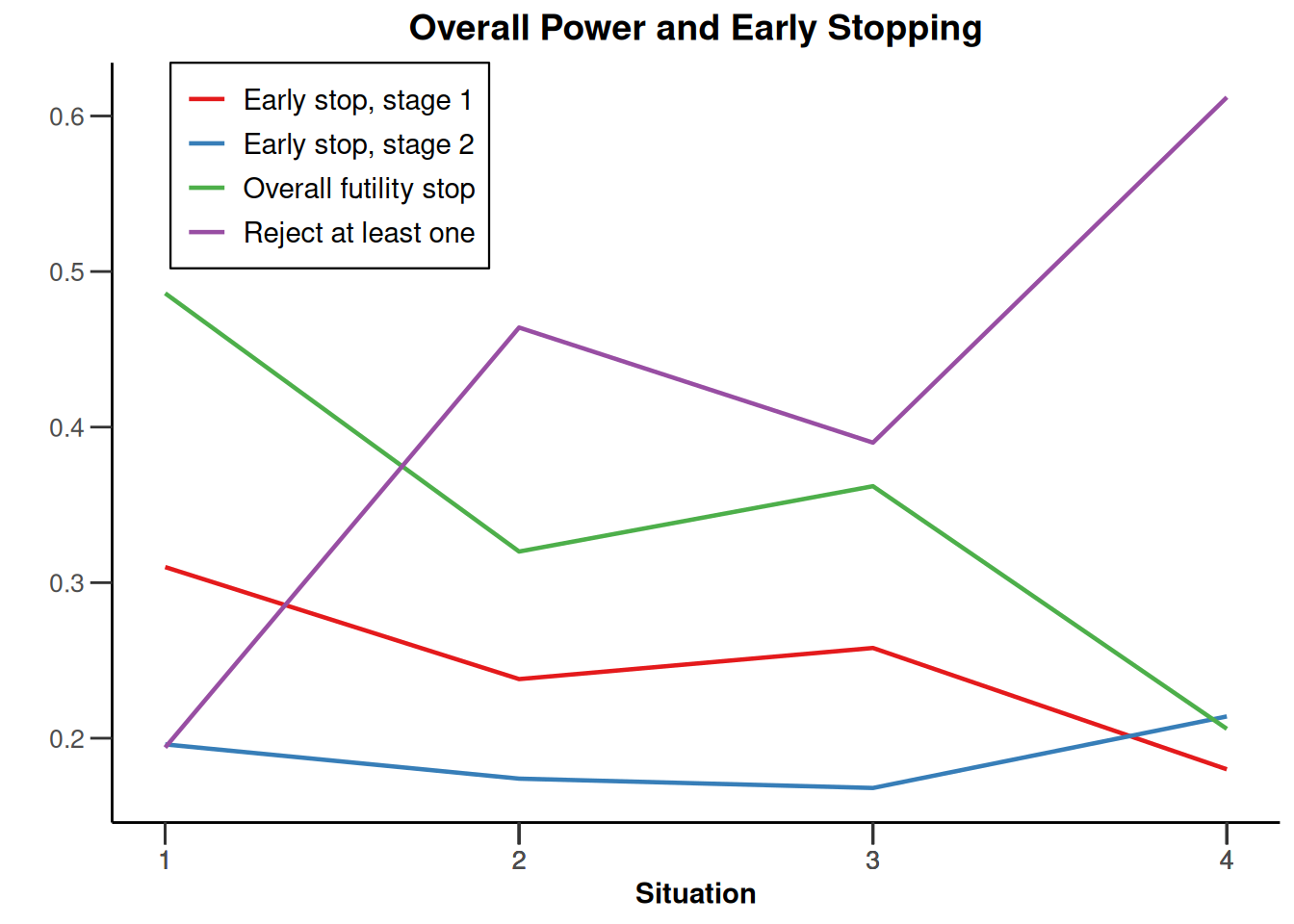
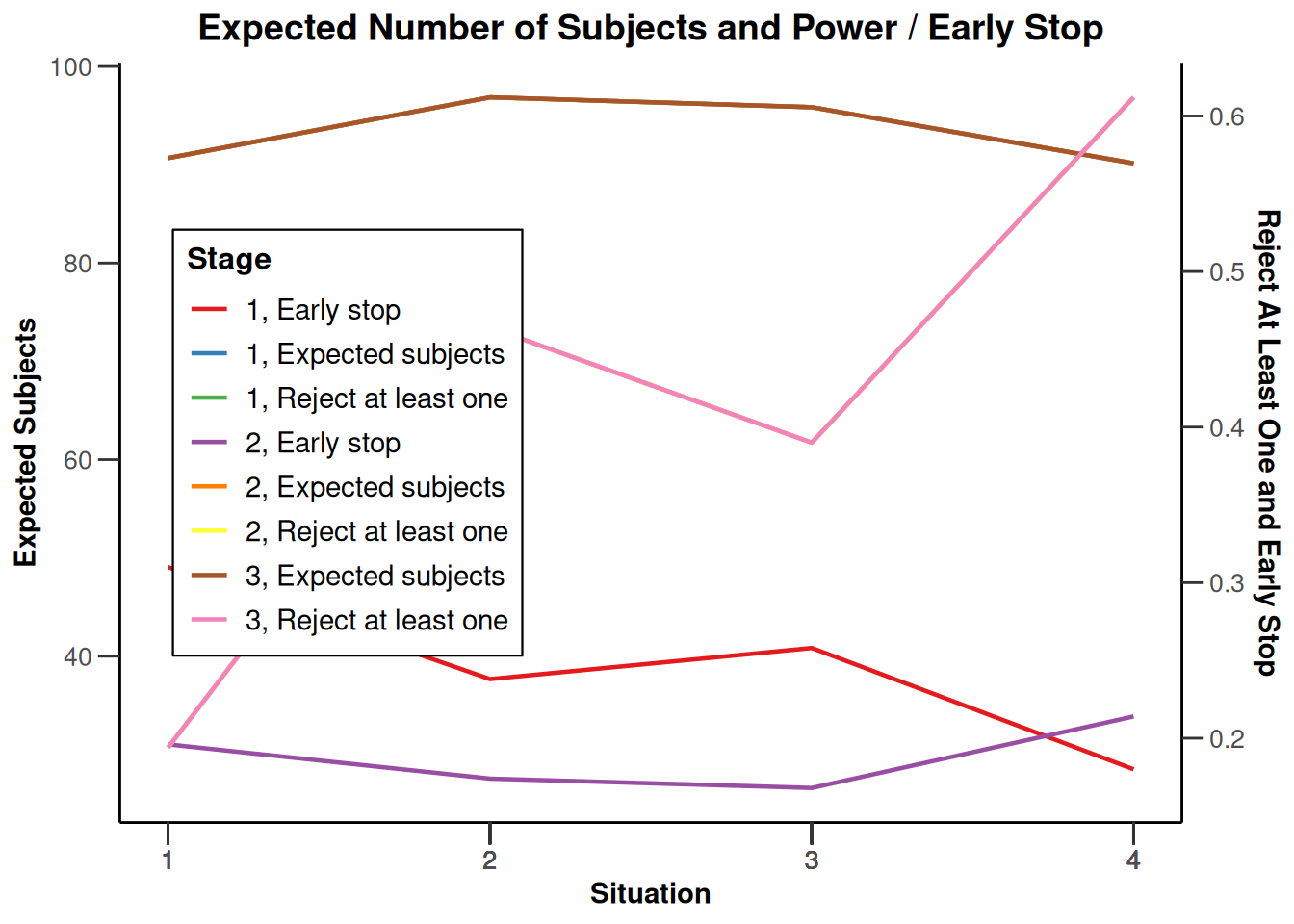
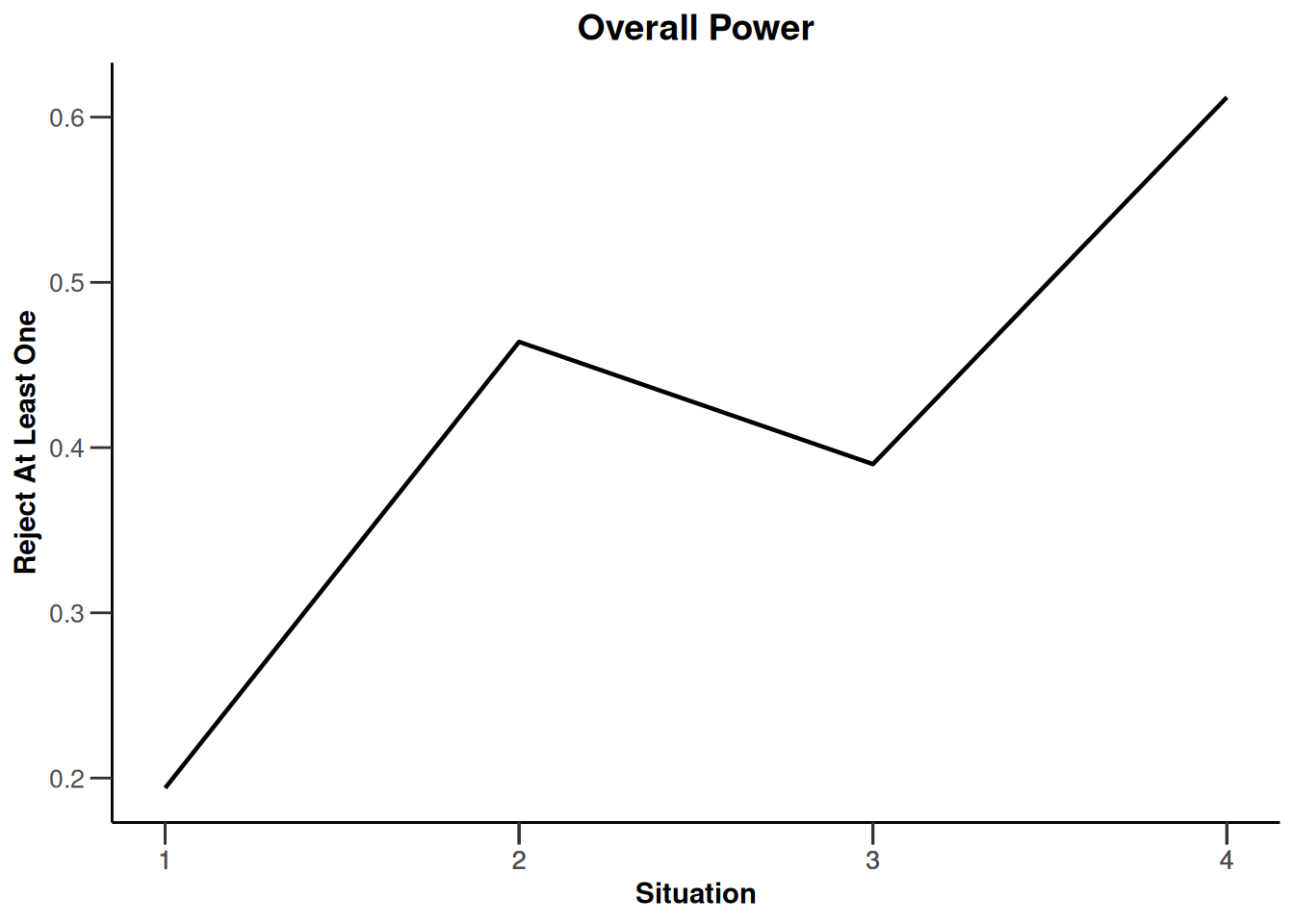
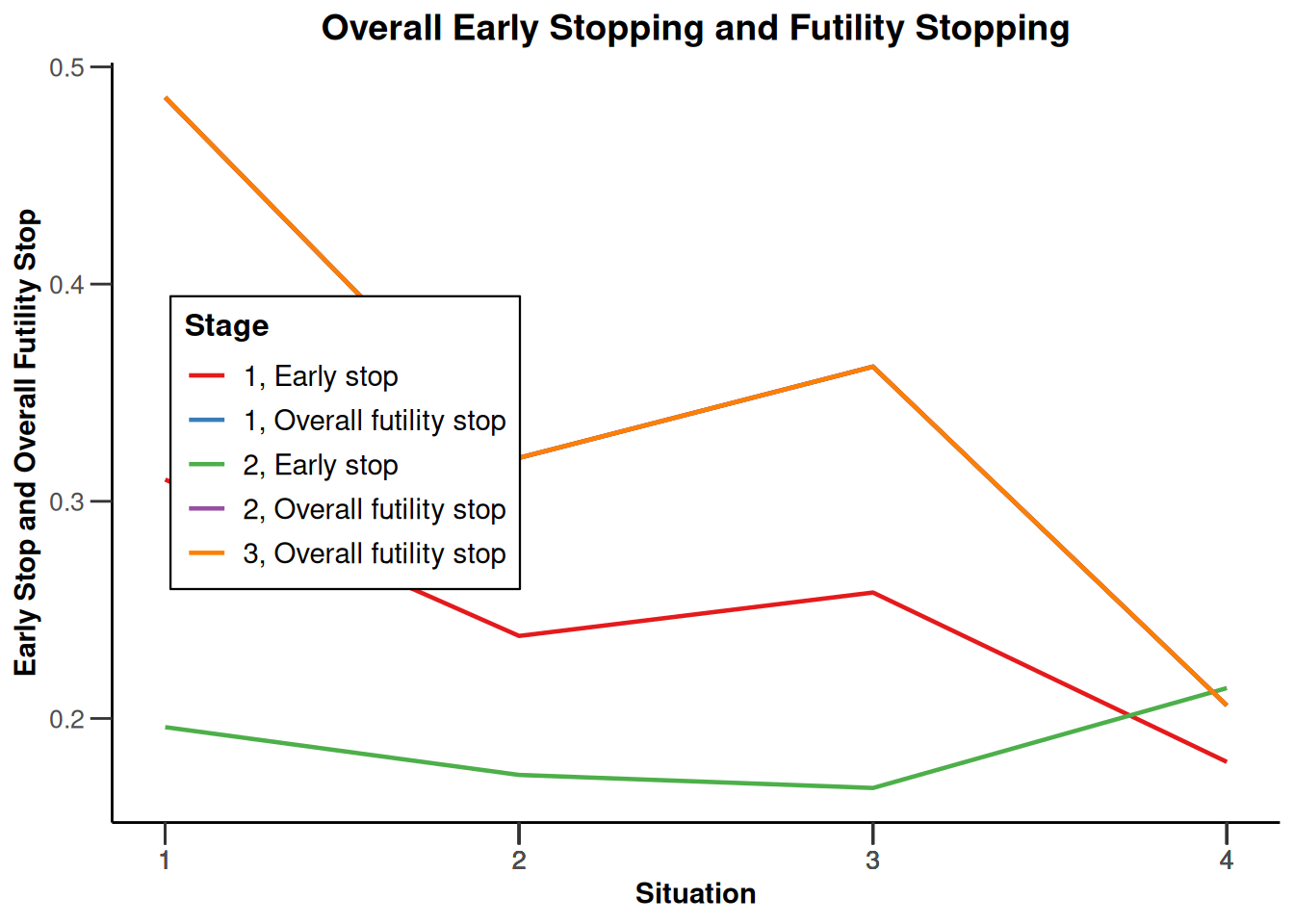
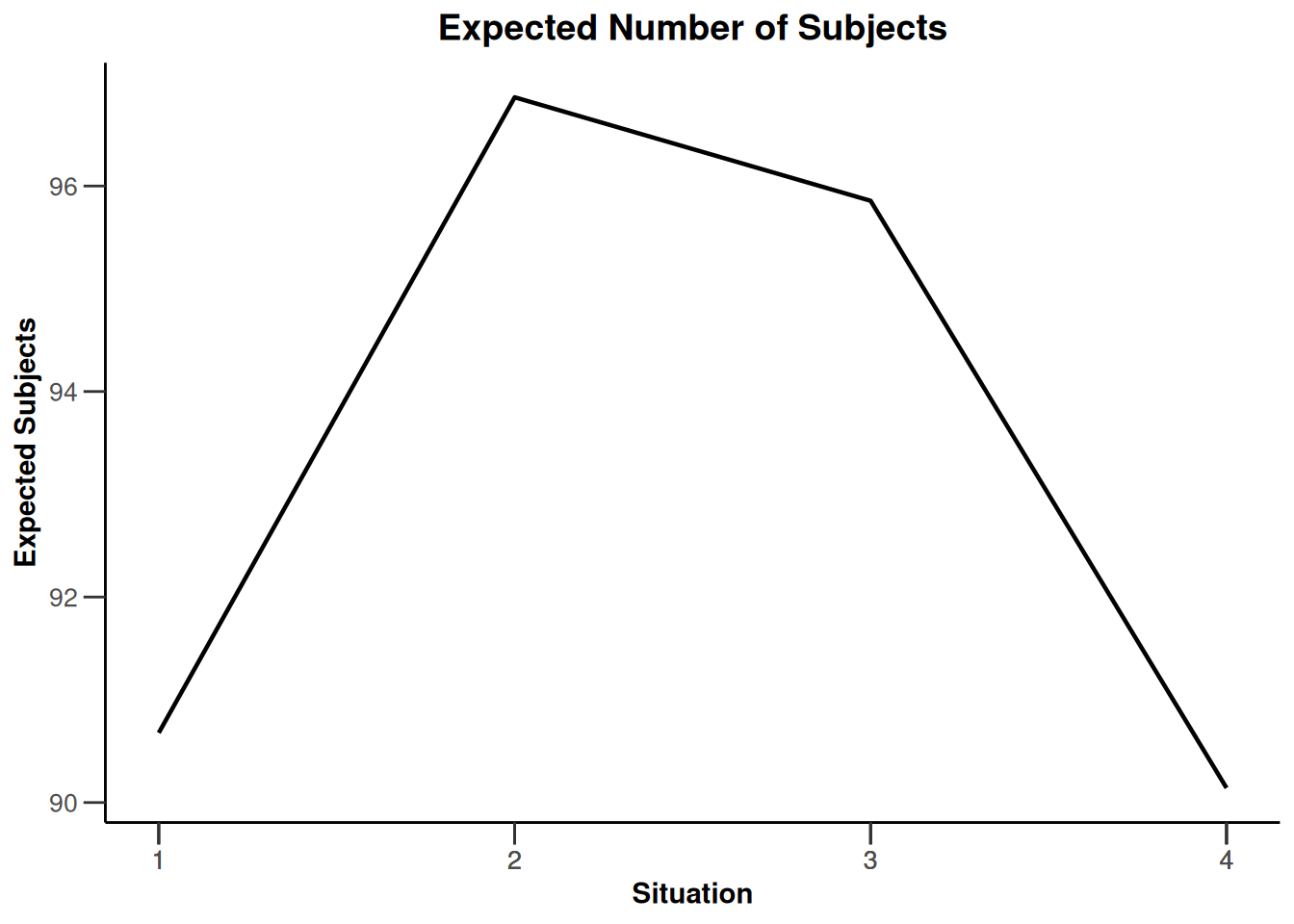
Simulation results enrichment - rates
design <- getDesignInverseNormal(
informationRates = c(0.2, 0.6, 1),
futilityBounds = c(-0.5, 0.5)
)
# Define effect list
subGroups <- c("S", "R")
prevalences <- c(0.4, 0.6)
piControl <- c(0.1, 0.4)
range1 <- piControl[1] + seq(0.0, 0.2, 0.1)
range2 <- piControl[2] + seq(0.0, 0.2, 0.1)
piTreatments <- c()
for (x1 in range1) {
for (x2 in range2) {
piTreatments <- c(piTreatments, x1, x2)
}
}
el <- list(
subGroups = subGroups,
prevalences = prevalences,
piControl = piControl,
piTreatments = matrix(piTreatments,
byrow = TRUE, ncol = 2
)
)
# Perform simulation
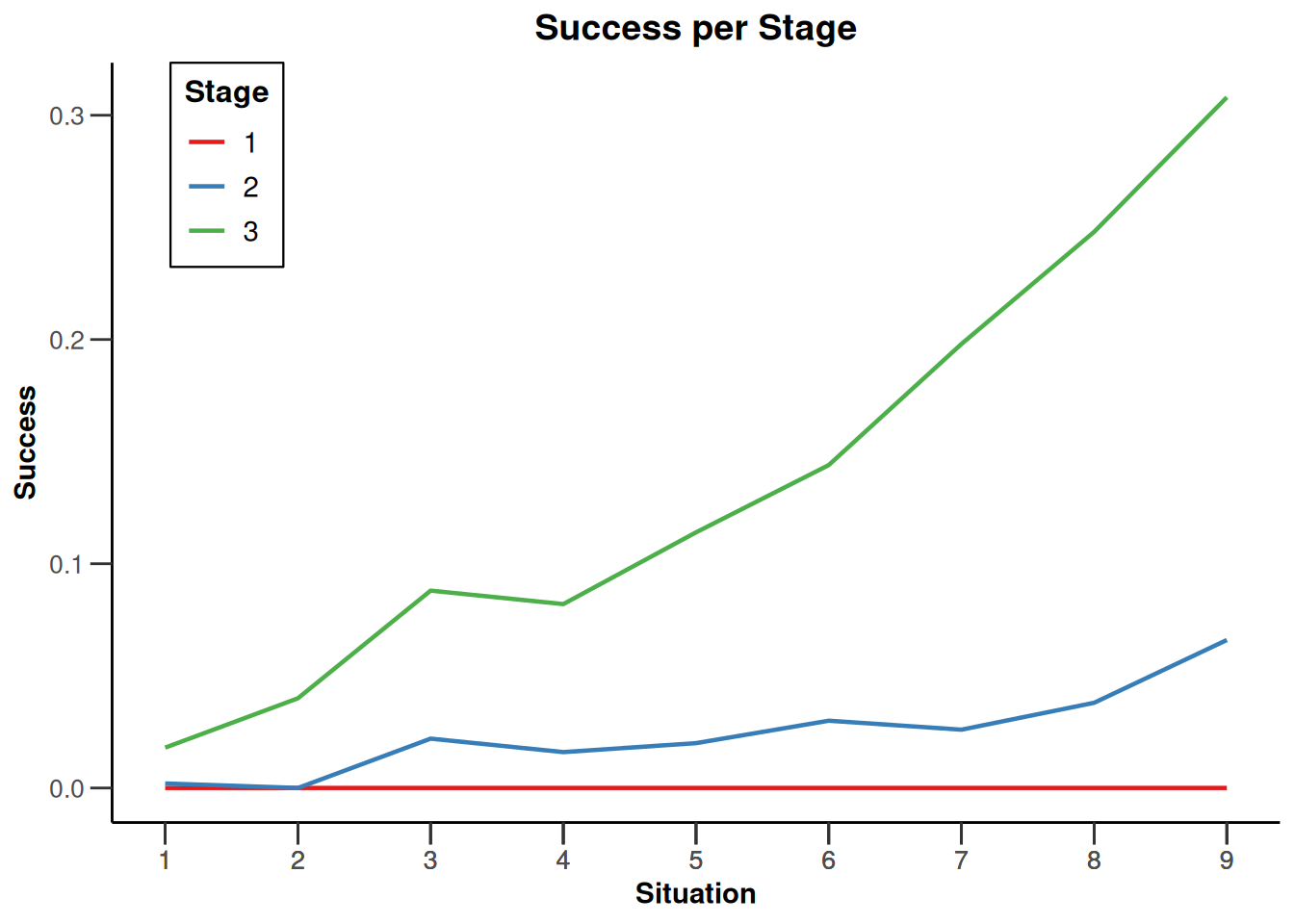
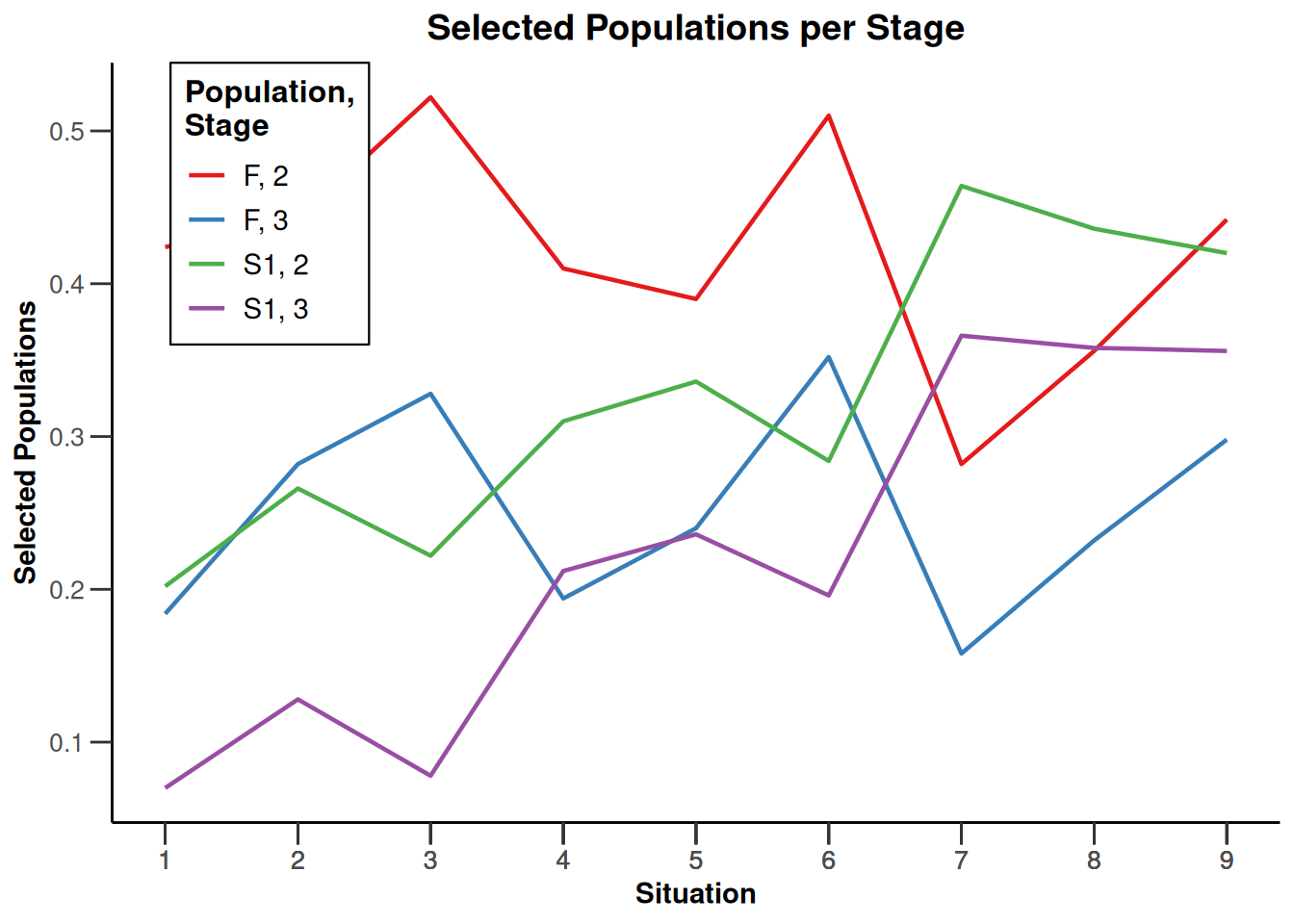
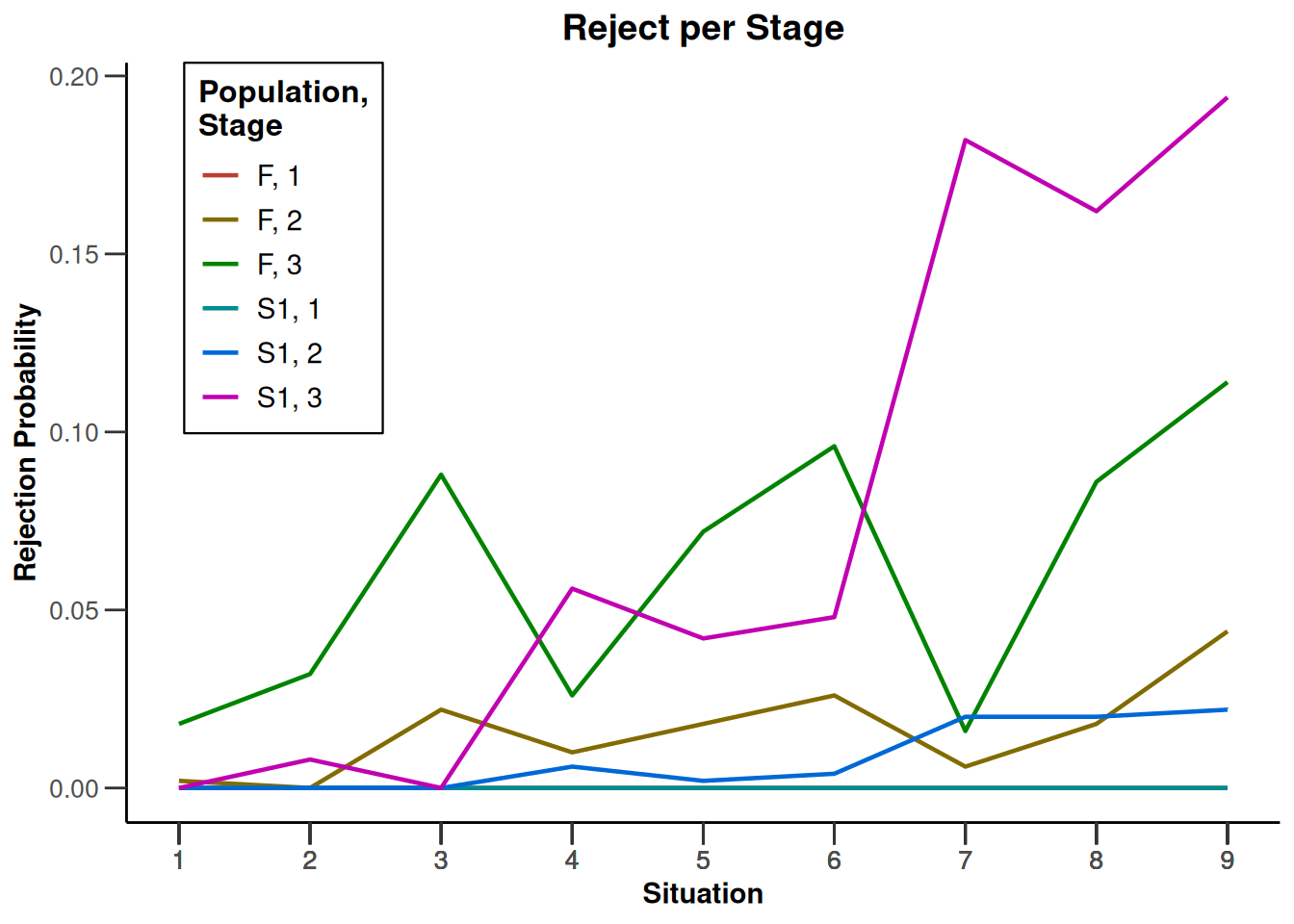
design |>
getSimulationEnrichmentRates(
plannedSubjects = c(10, 30, 50),
effectList = el,
maxNumberOfIterations = 500,
seed = 1234567890
) |>
plot(type = "all", grid = 0)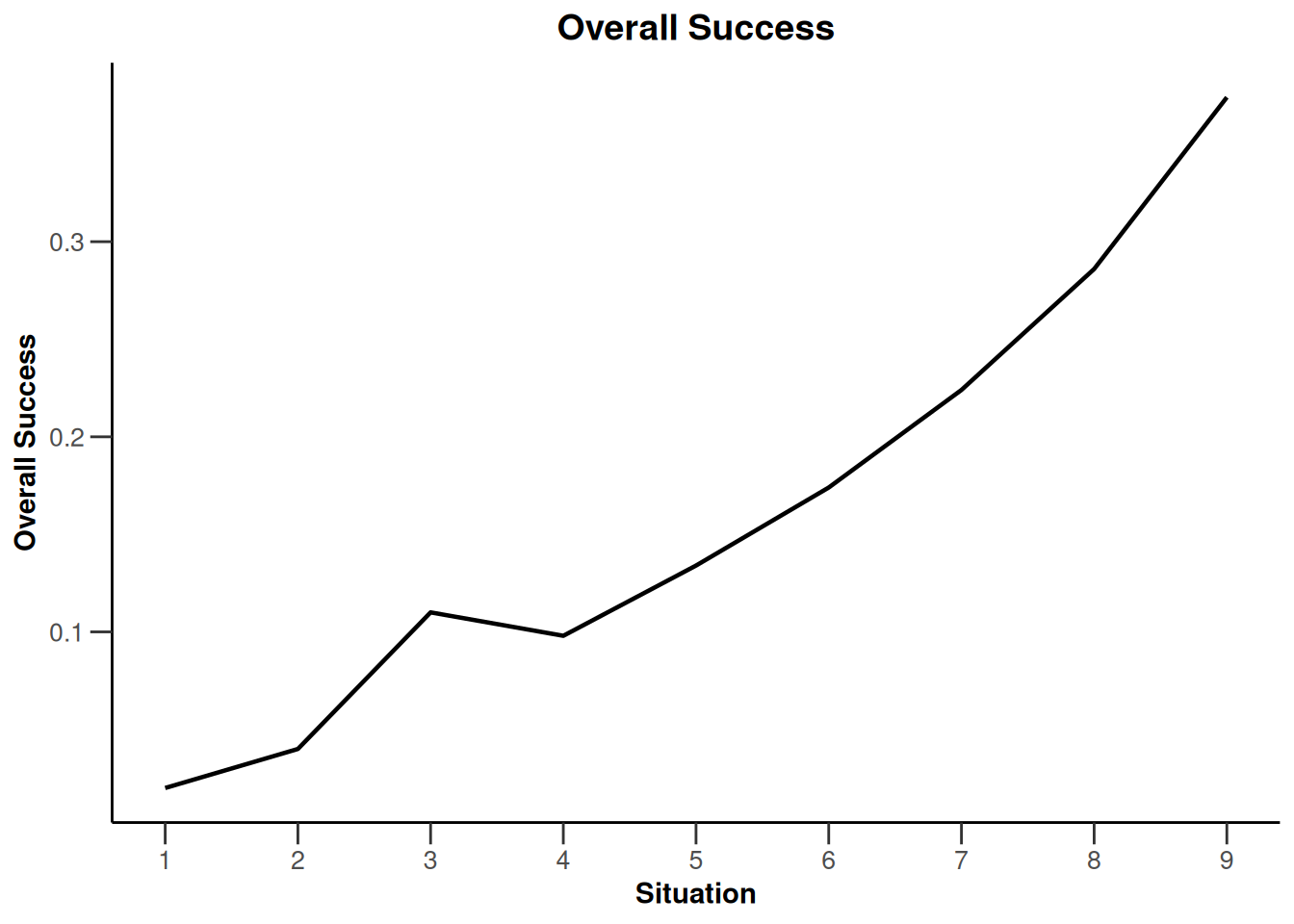
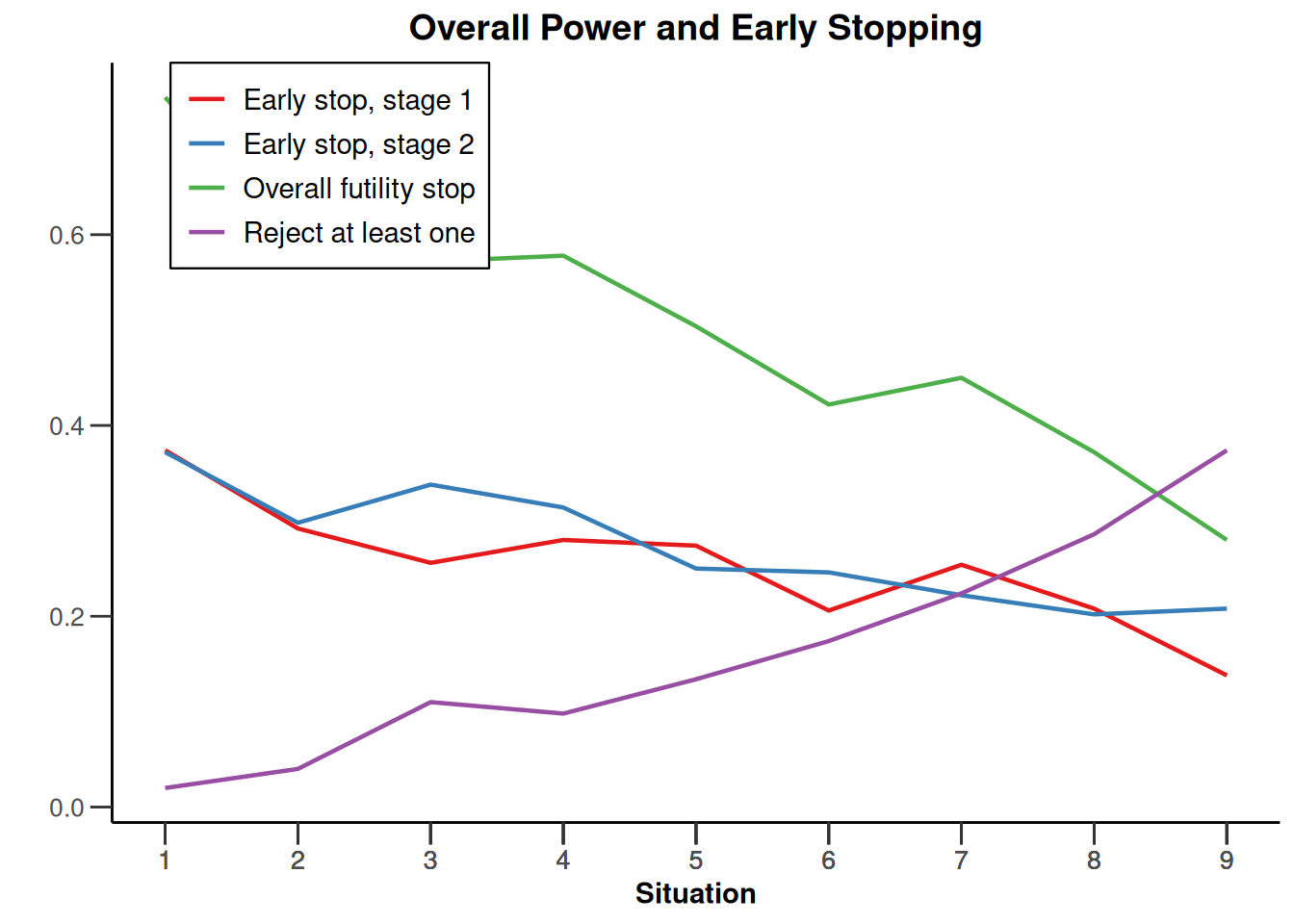
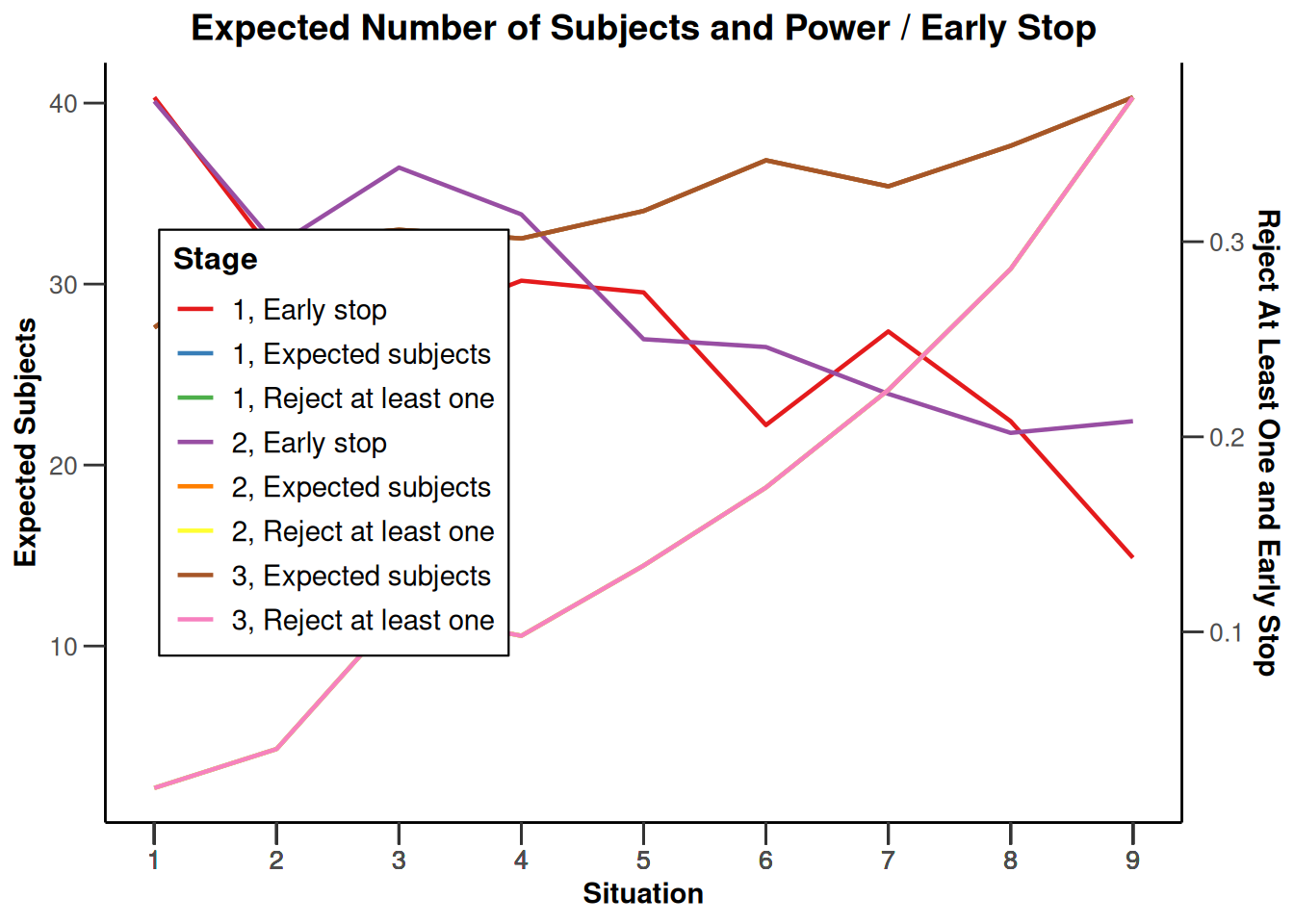
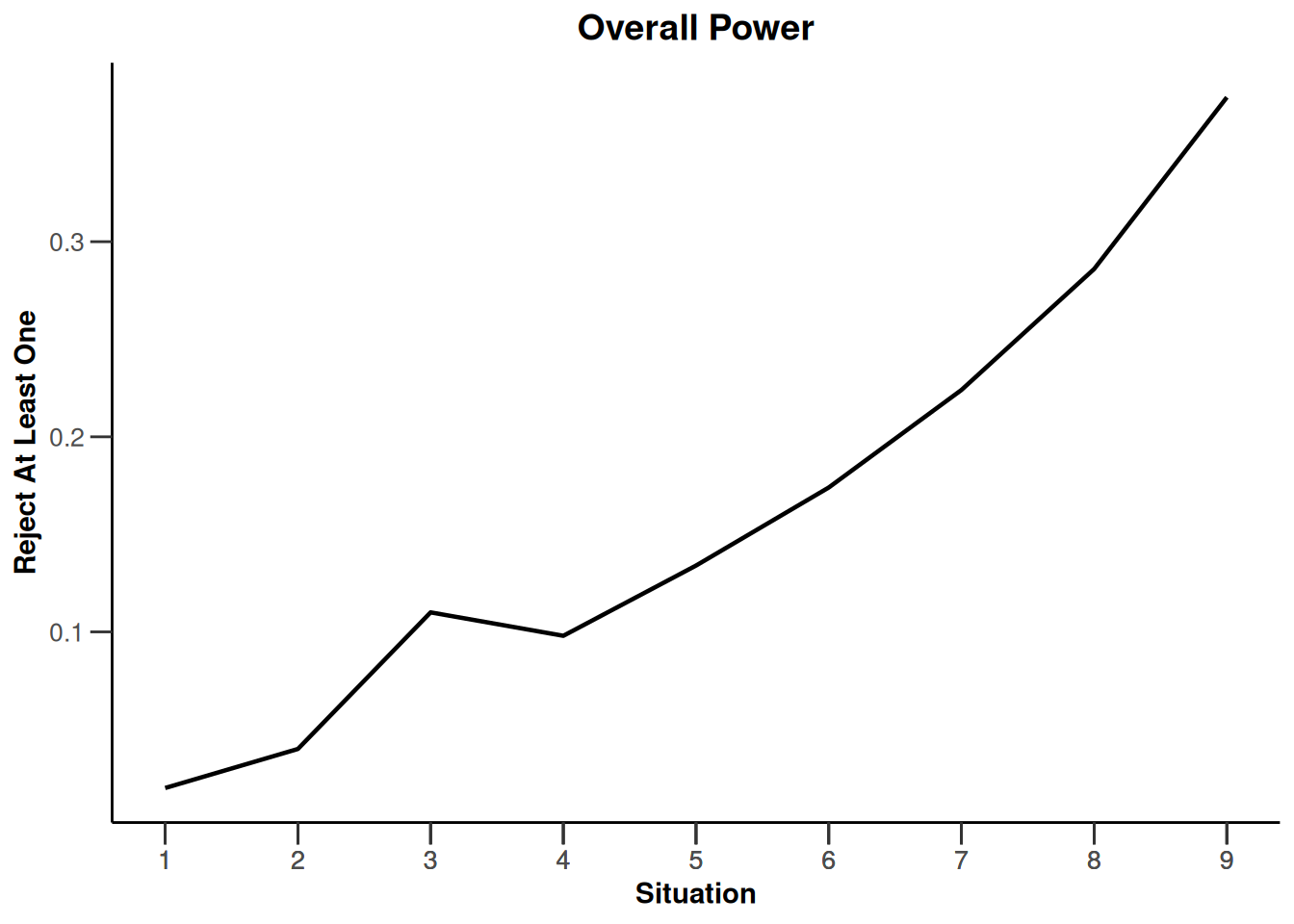
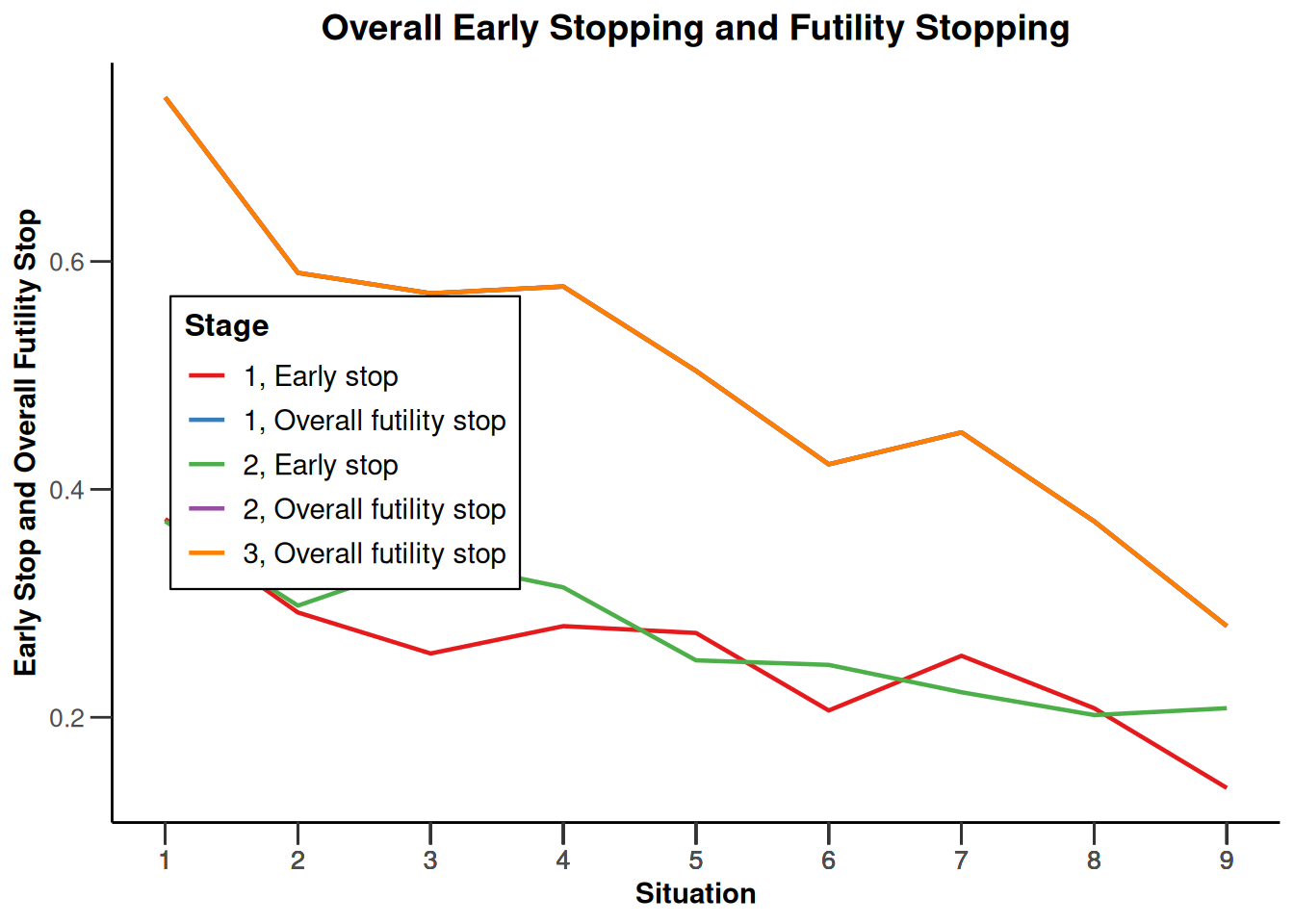
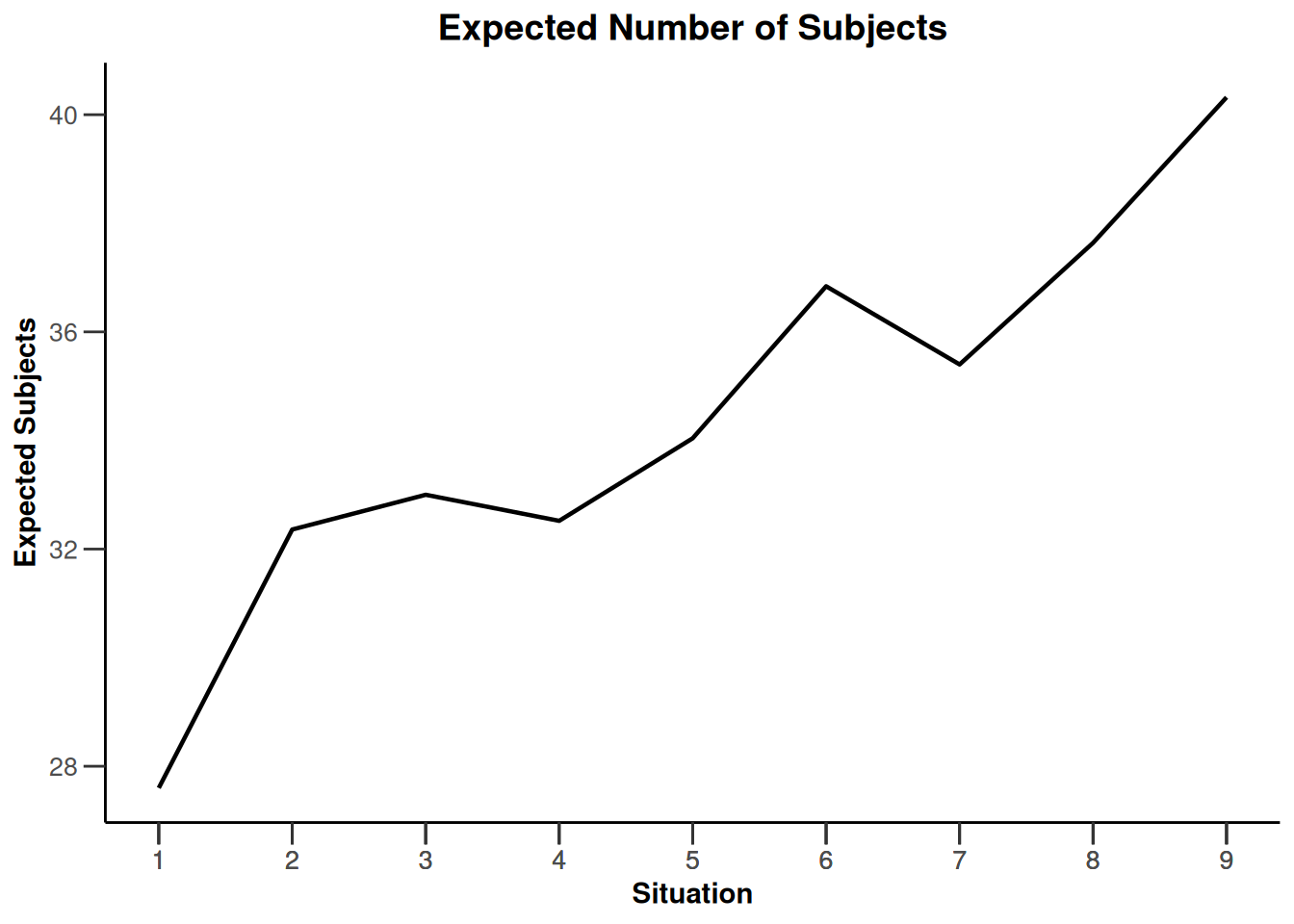
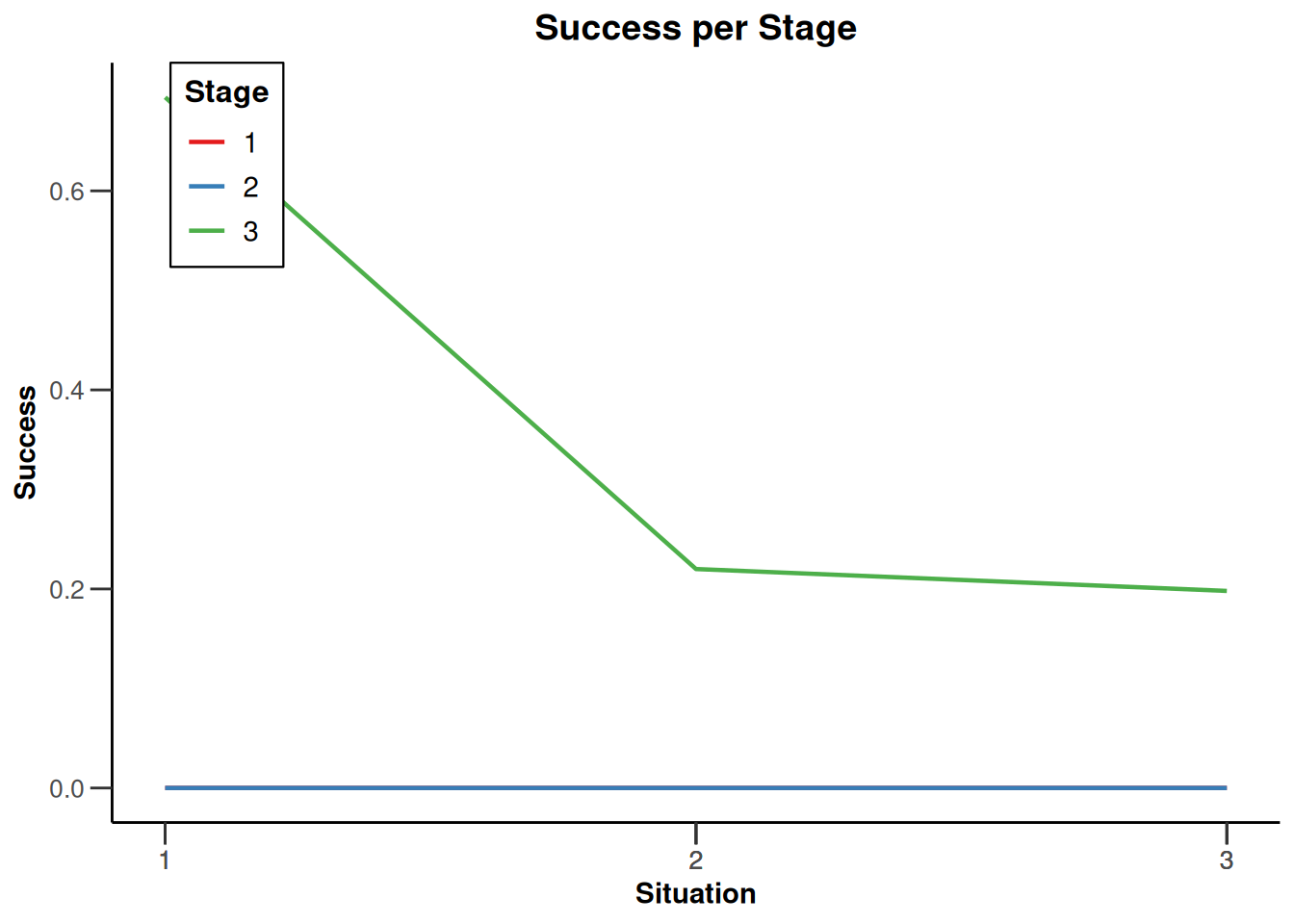
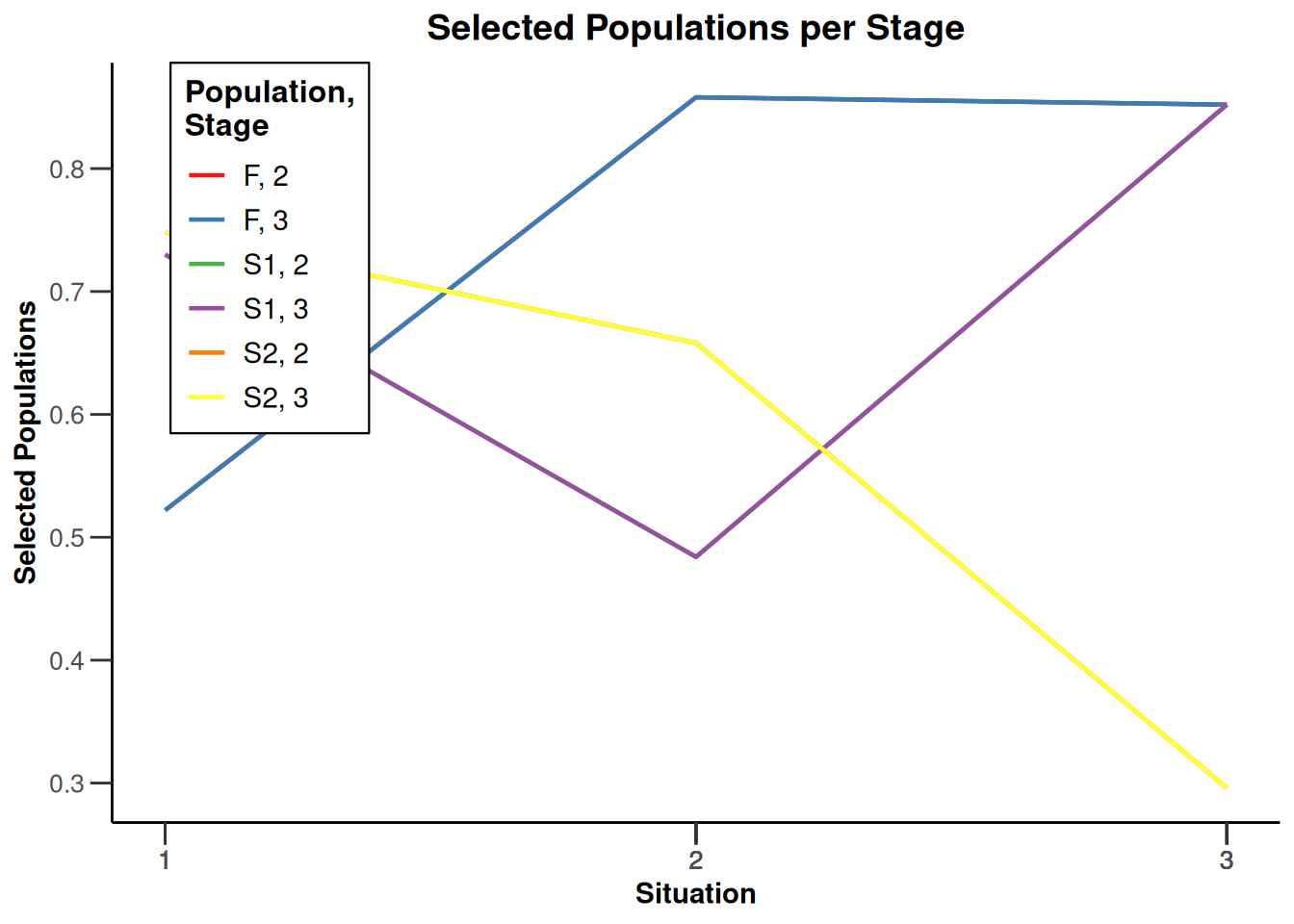
Simulation results enrichment - survival
# Define subgroups and their prevalences
subGroups <- c("S1", "S2", "S12", "R") # fixed names!
prevalences <- c(0.2, 0.3, 0.4, 0.1)
piControls <- c(0.2, 0.4, 0.15, 0.3)
effect <- c(-0.05, -0.02, -0.10, -0.10)
piTreatments <- piControls + effect
hr <- log(1 - piTreatments) / log(1 - piControls)
# Define effect list
el <- list(
subGroups = subGroups, prevalences = prevalences,
piControls = piControls, hazardRatios = matrix(rep(hr, 3), nrow = 3)
)
# Perform simulation
getDesignInverseNormal(
typeOfDesign = "noEarlyEfficacy") |>
getSimulationEnrichmentSurvival(
effectList = el,
typeOfSelection = "rbest",
rValue = 2,
intersectionTest = "Simes",
plannedEvents = c(30, 80, 120),
maxNumberOfIterations = 500,
directionUpper = FALSE
) |>
plot(type = "all", grid = 0)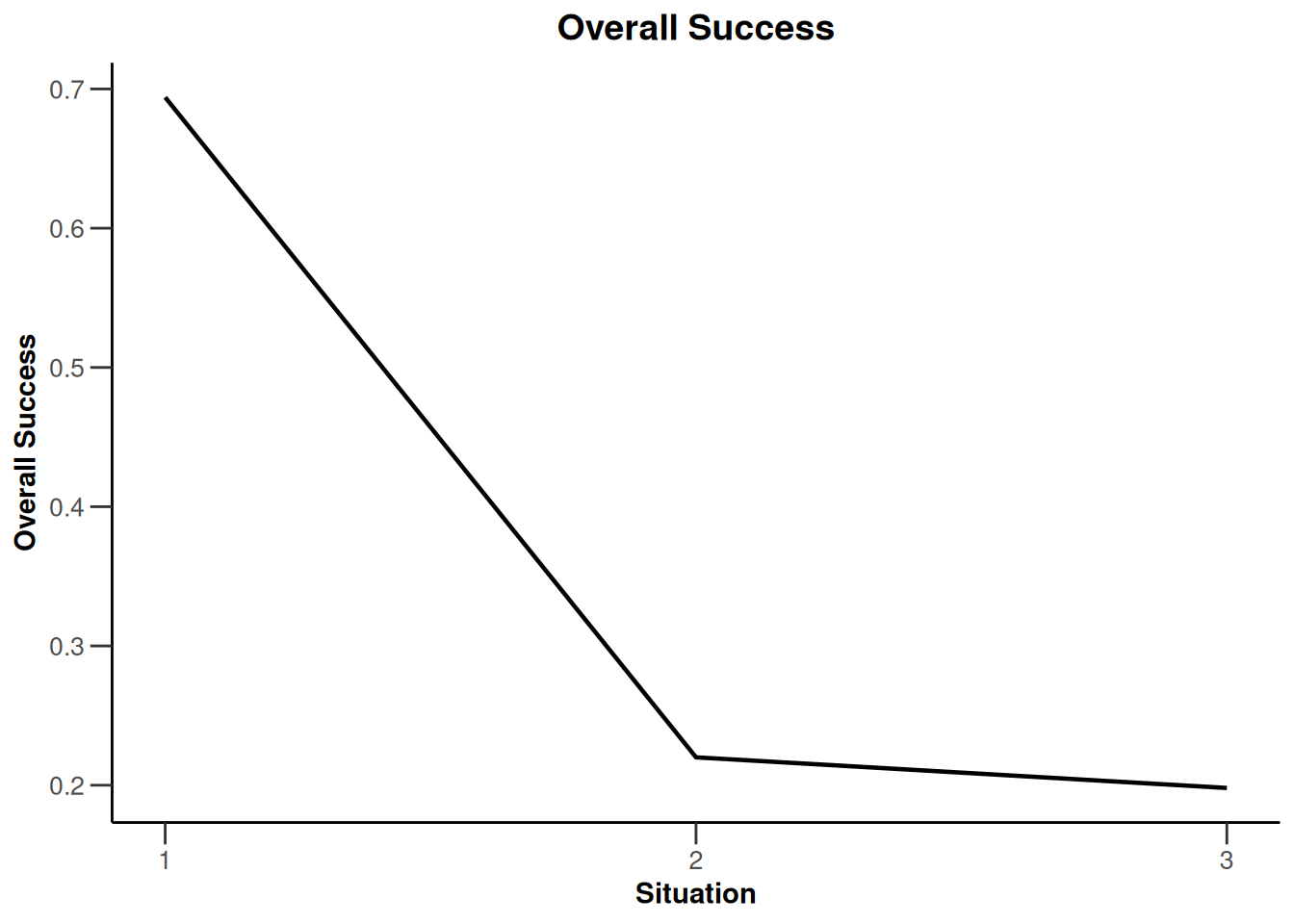
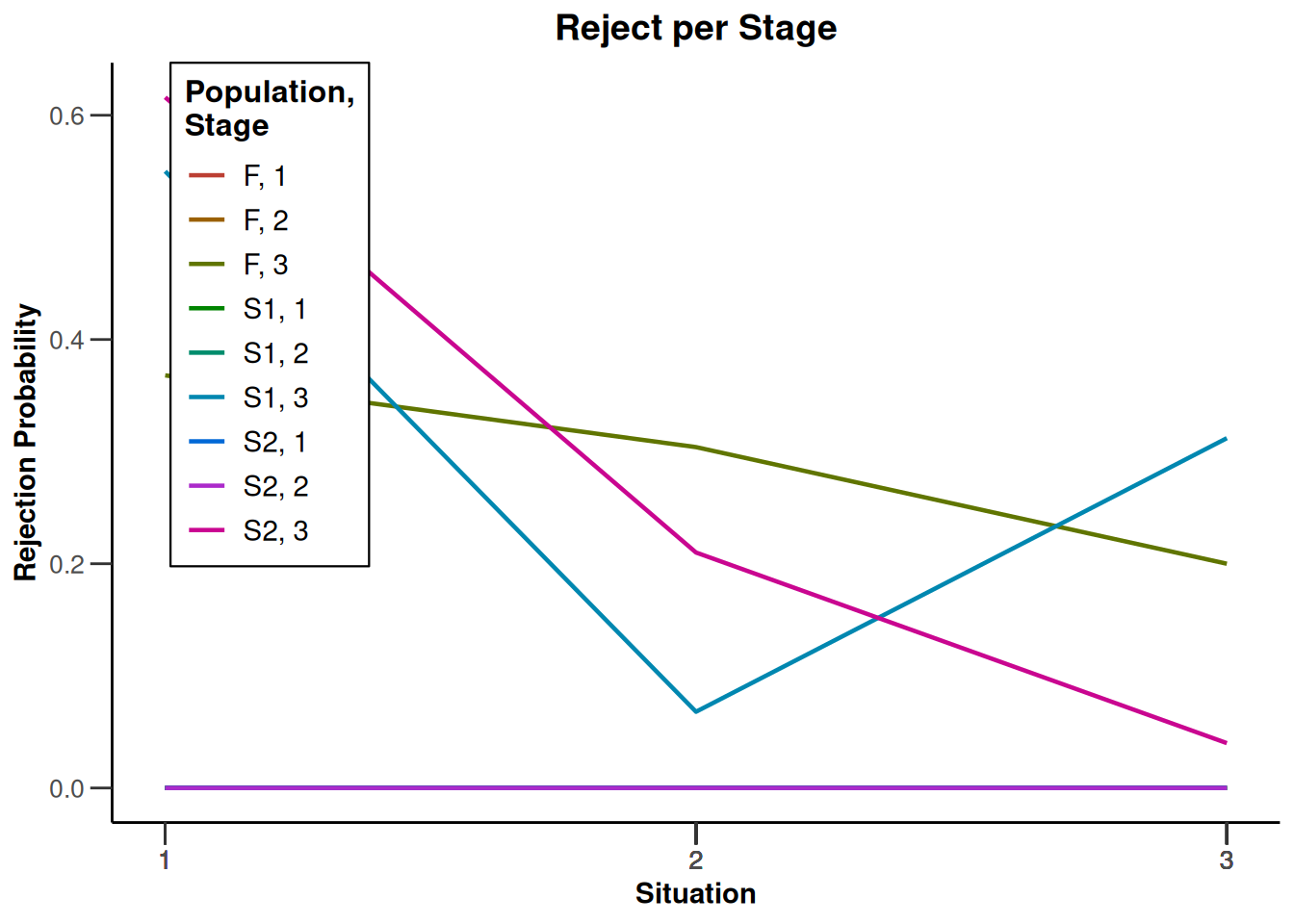
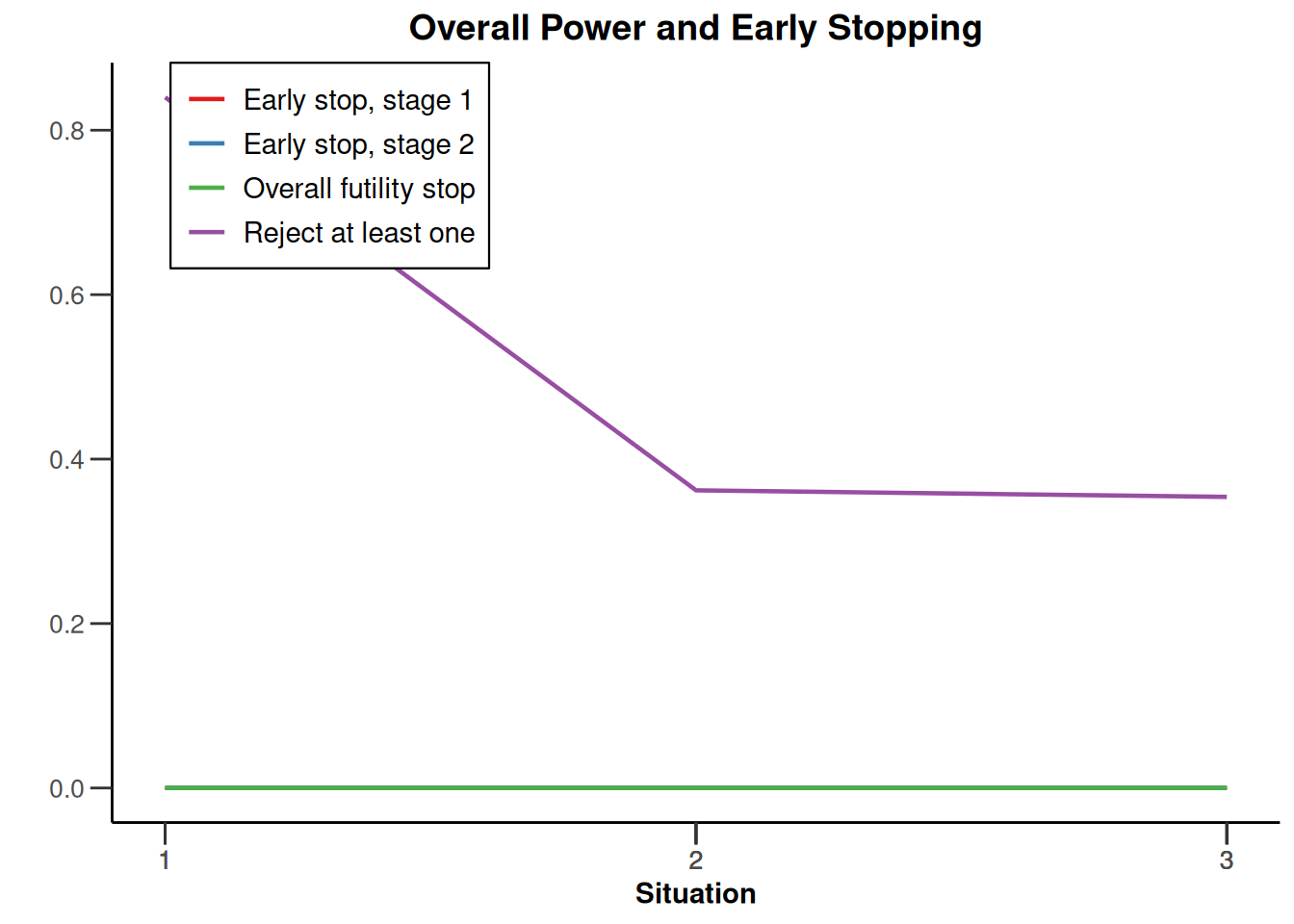
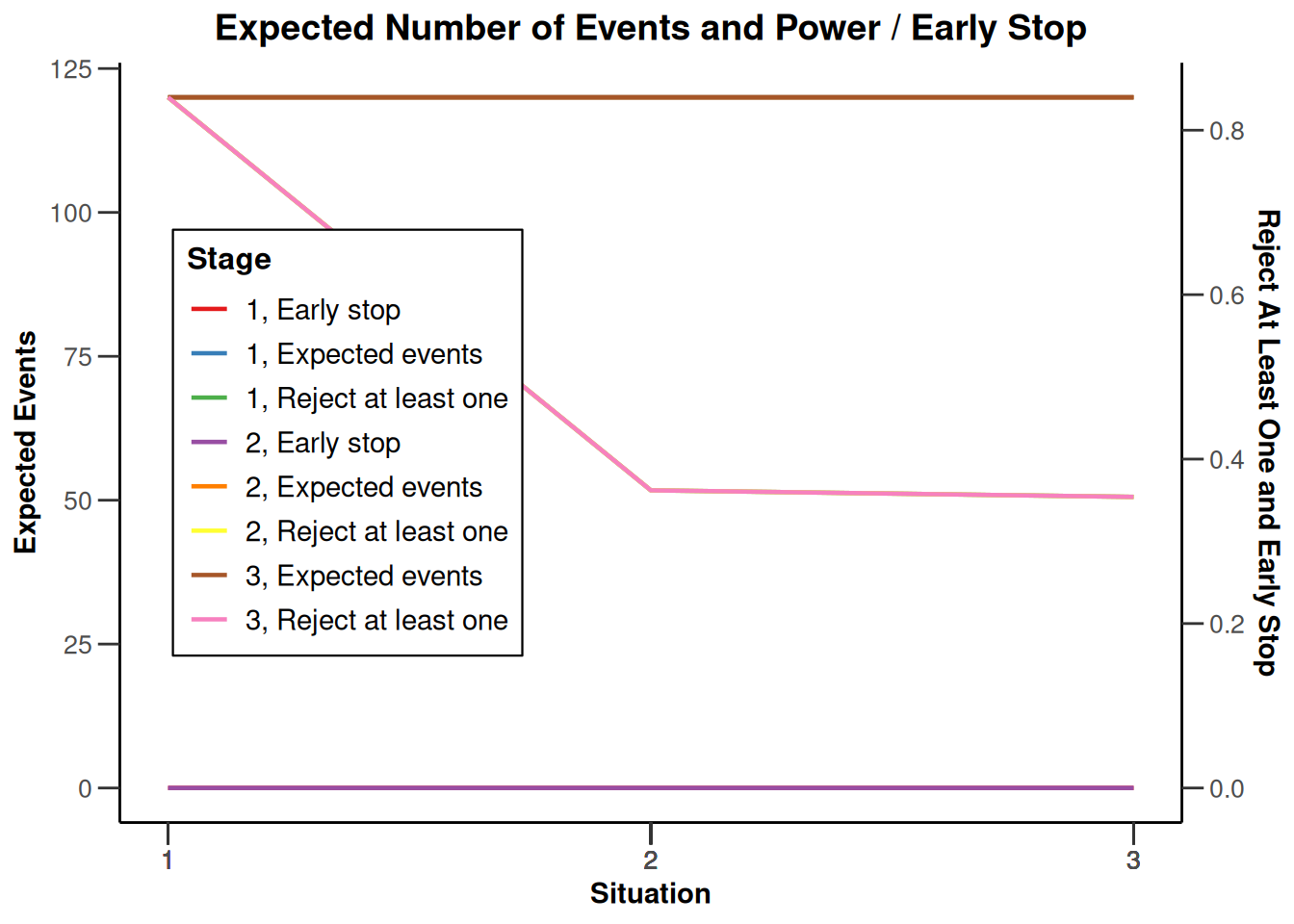
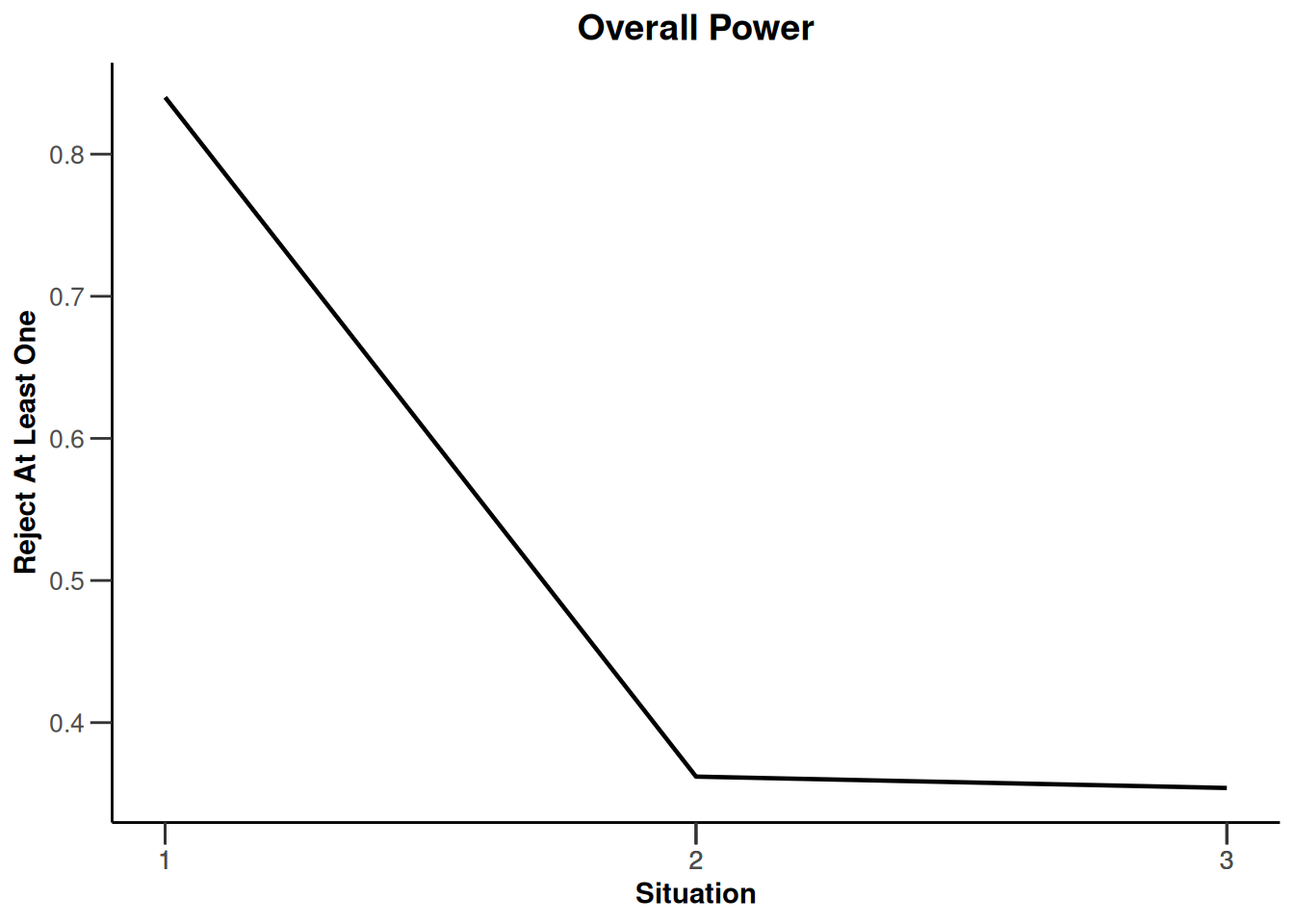
System: rpact 4.3.0, R version 4.5.2 (2025-10-31), platform: x86_64-pc-linux-gnu
To cite R in publications use:
R Core Team (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
To cite package ‘rpact’ in publications use:
Wassmer G, Pahlke F (2025). rpact: Confirmatory Adaptive Clinical Trial Design and Analysis. doi:10.32614/CRAN.package.rpact https://doi.org/10.32614/CRAN.package.rpact, R package version 4.2.0, https://cran.r-project.org/package=rpact.