library(rpact)
packageVersion("rpact") # version should be version 3.0 or laterHow to Create Admirable Plots with rpact
Utilities
Sample size
Power simulation
Preparation
First, load the rpact package
[1] '4.3.0'Design plots
One-sided design with futility bounds
design <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(0, 0.1)
)
design |> plot(type = 1)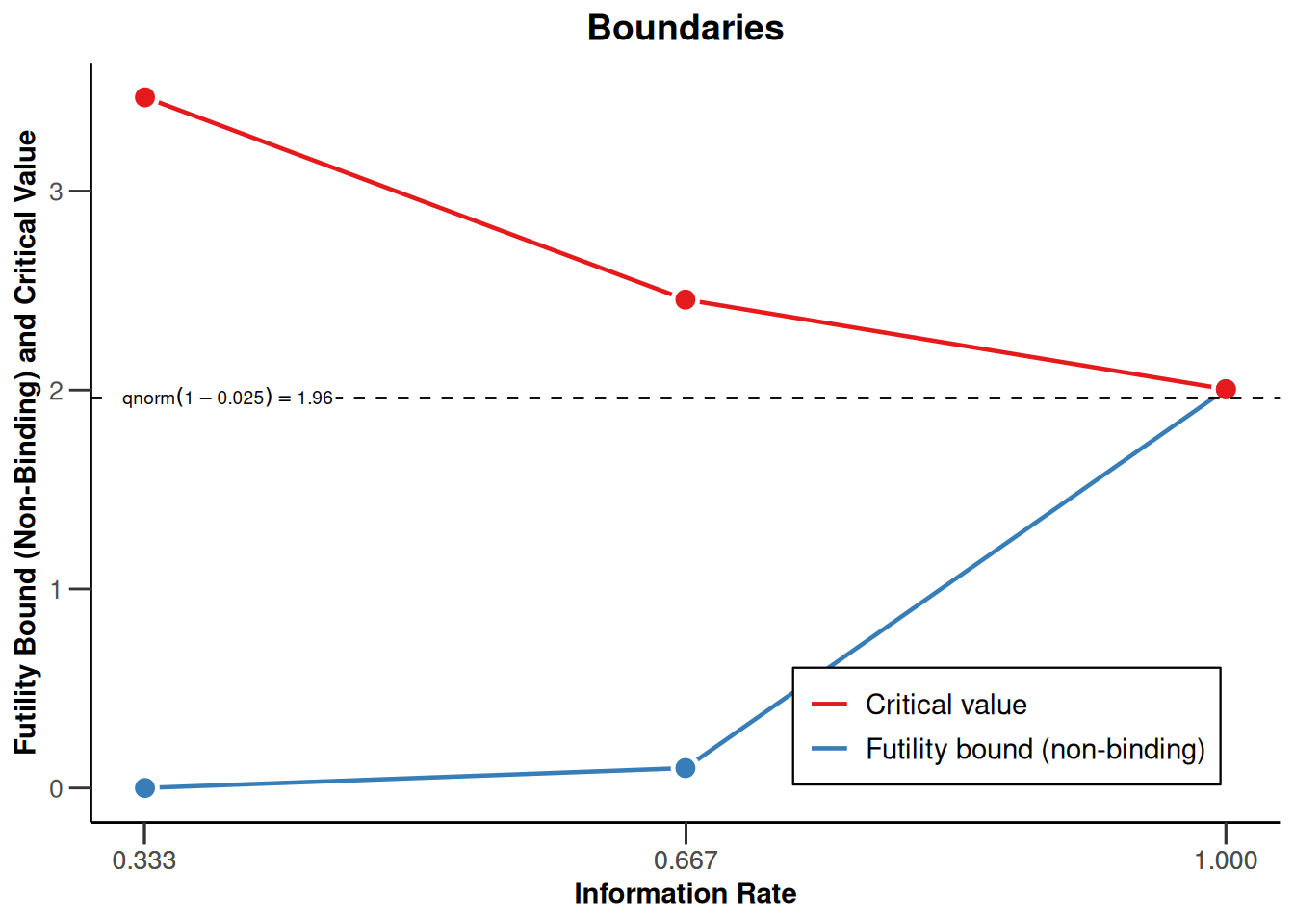
design |> plot(type = 5, nMax = 10)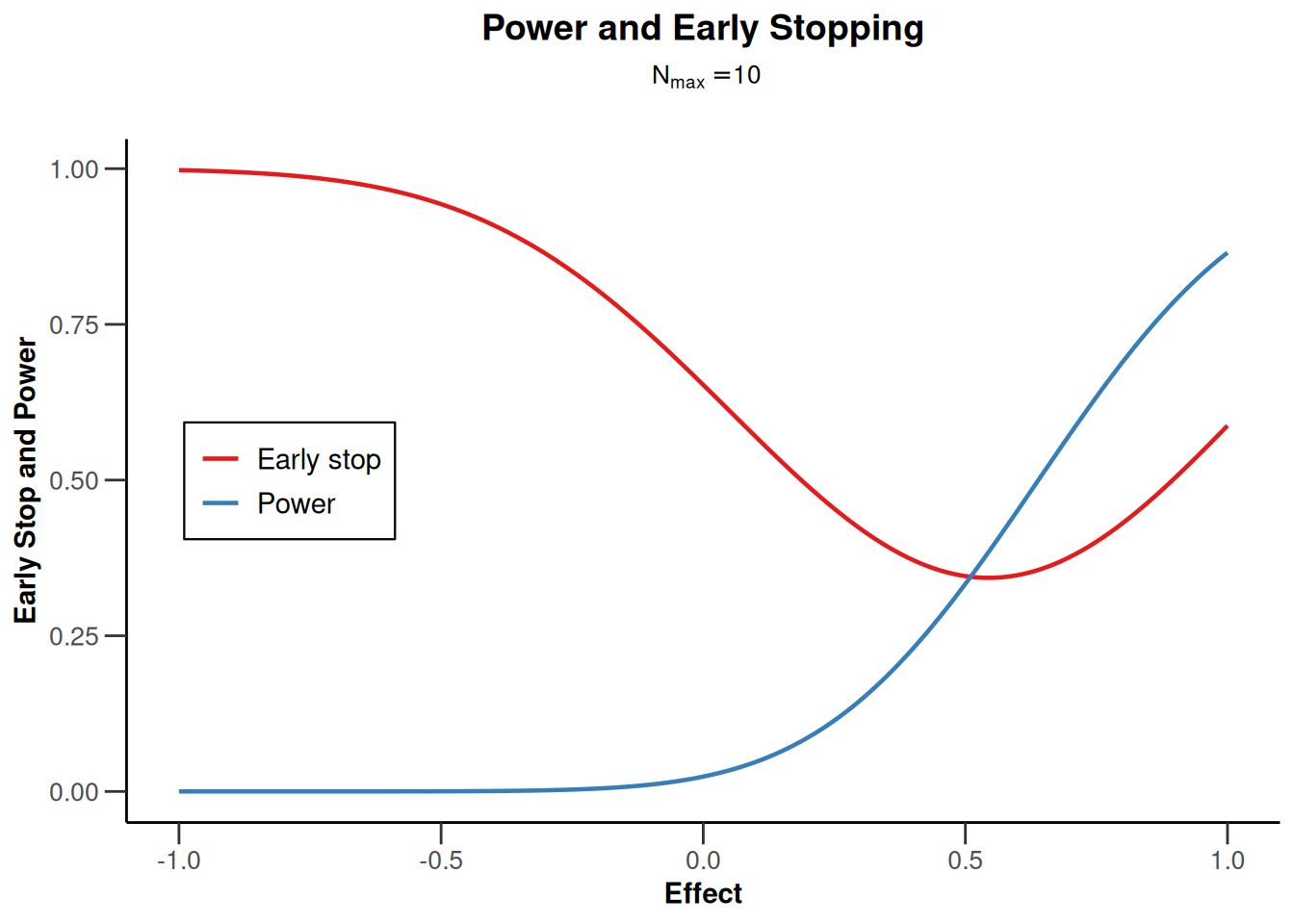
design |> plot(type = 6, nMax = 10)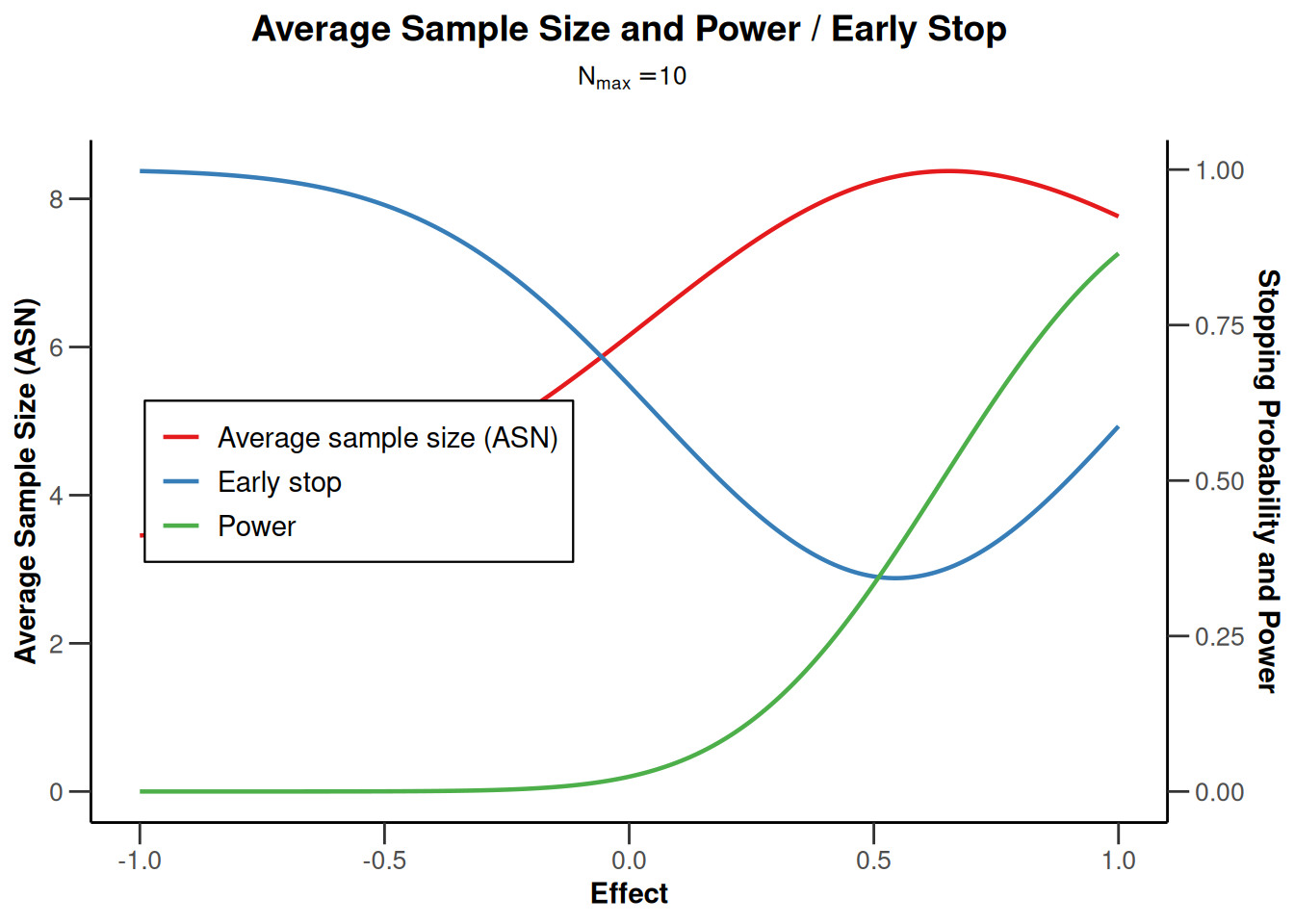
Two-sided design
design <- getDesignGroupSequential(
kMax = 4,
typeOfDesign = "OF",
sided = 2,
twoSidedPower = TRUE
)
design |> plot(type = 1)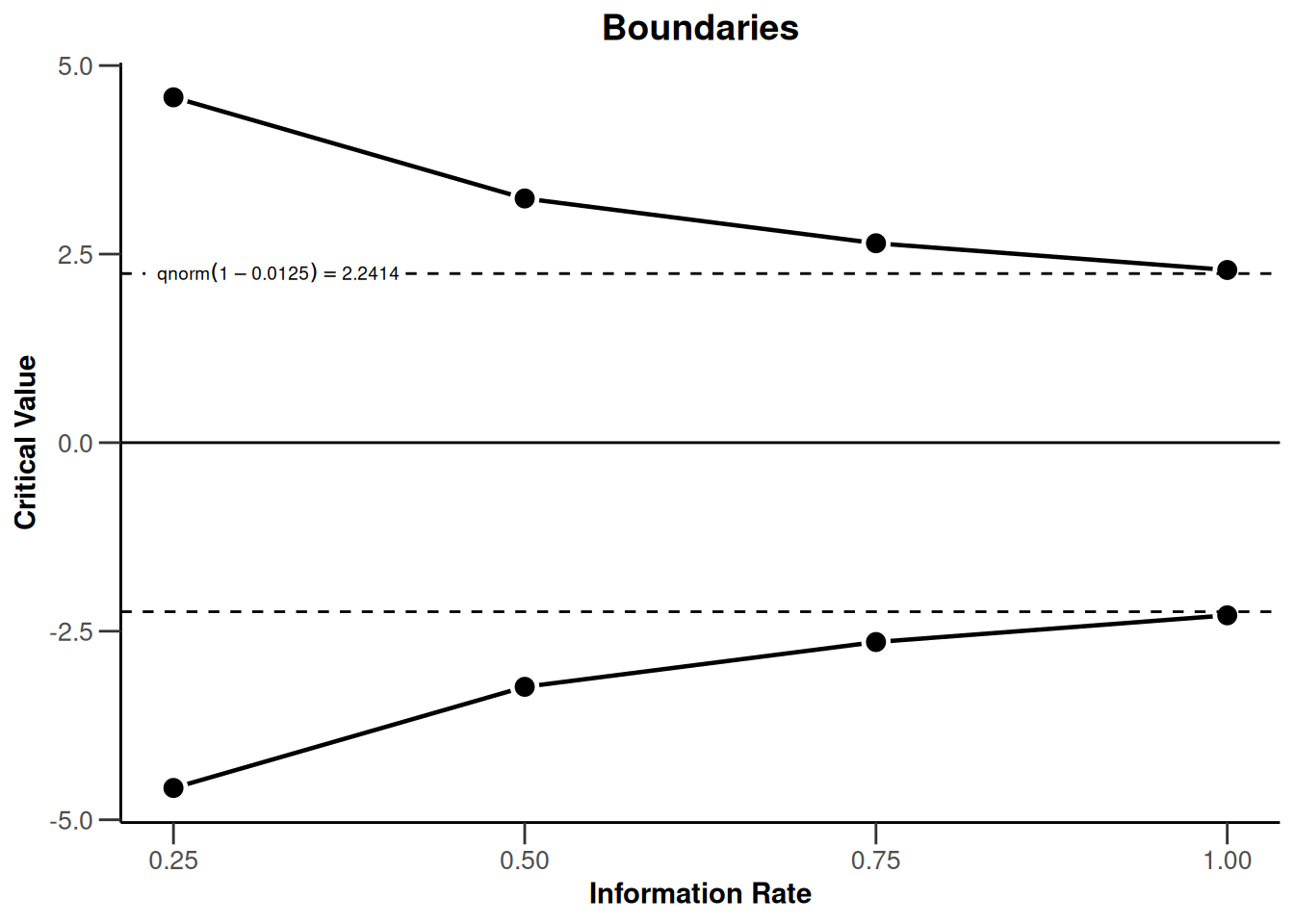
design |> plot(type = 5, nMax = 10)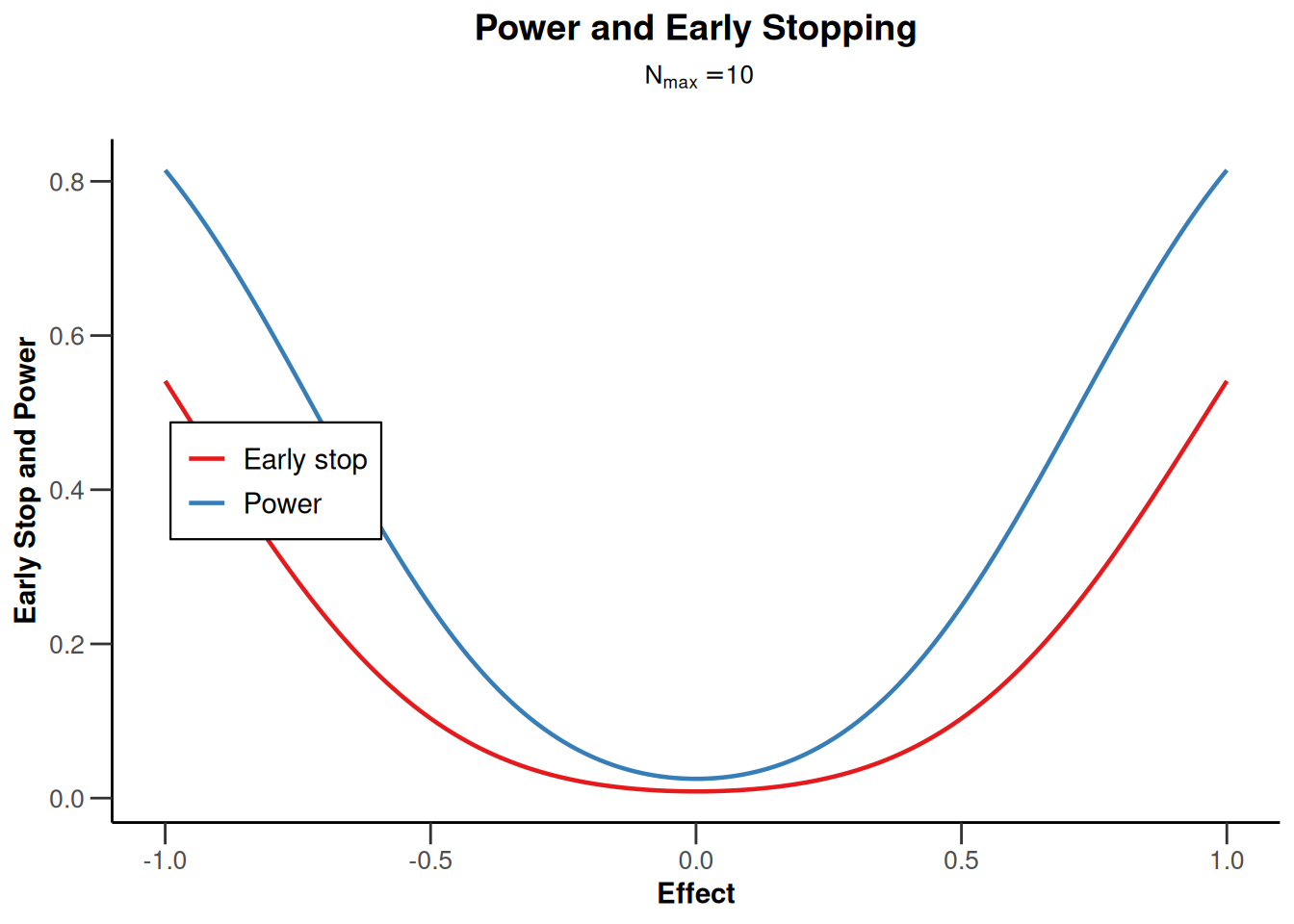
design |> plot(type = 6, nMax = 10)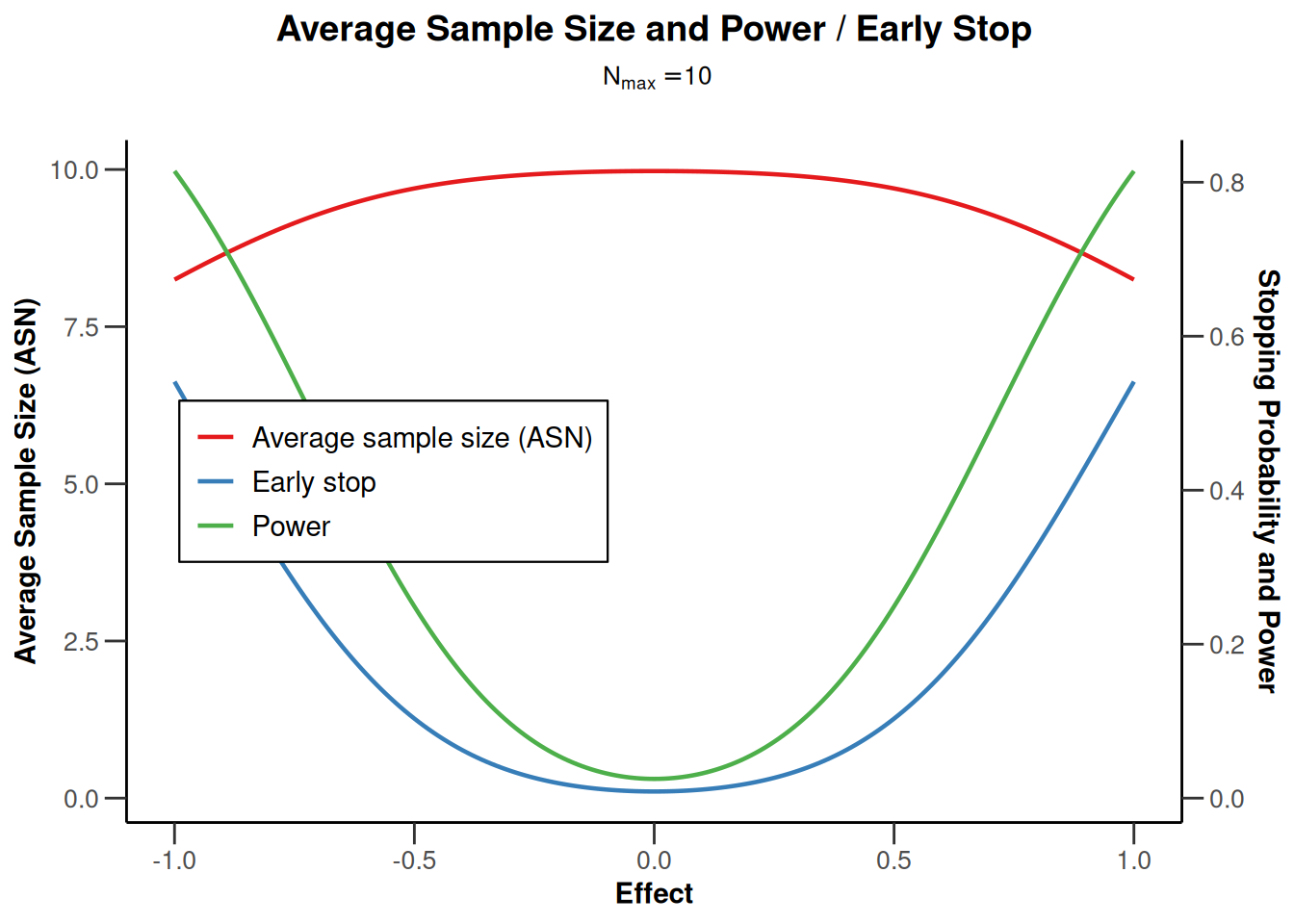
Two-sided design with futility bound
design <- getDesignGroupSequential(
beta = 0.05,
kMax = 4,
typeOfDesign = "asOF",
typeBetaSpending = "bsOF",
sided = 2
)
design |> plot(type = 1)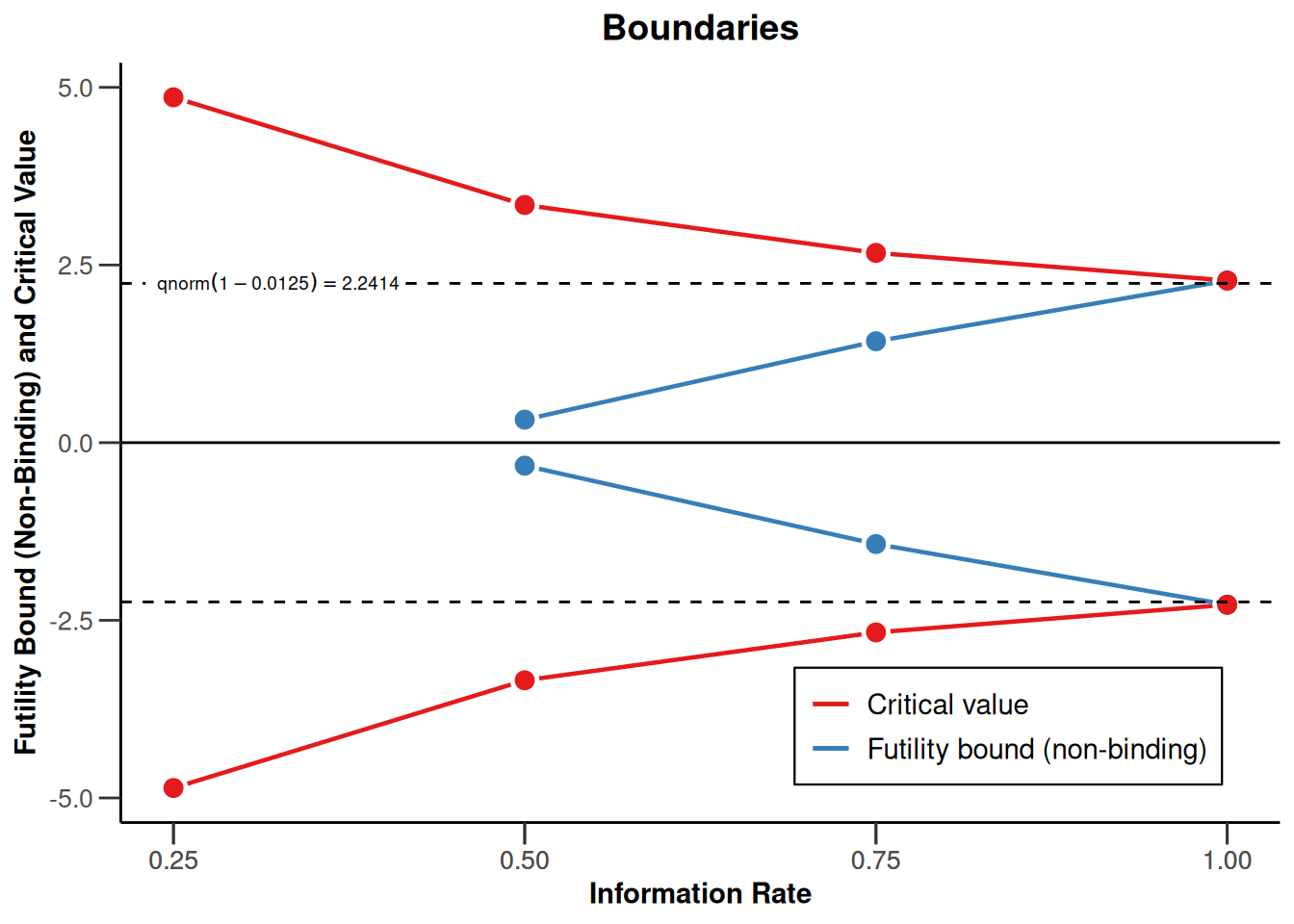
design |> plot(type = 5, nMax = 10)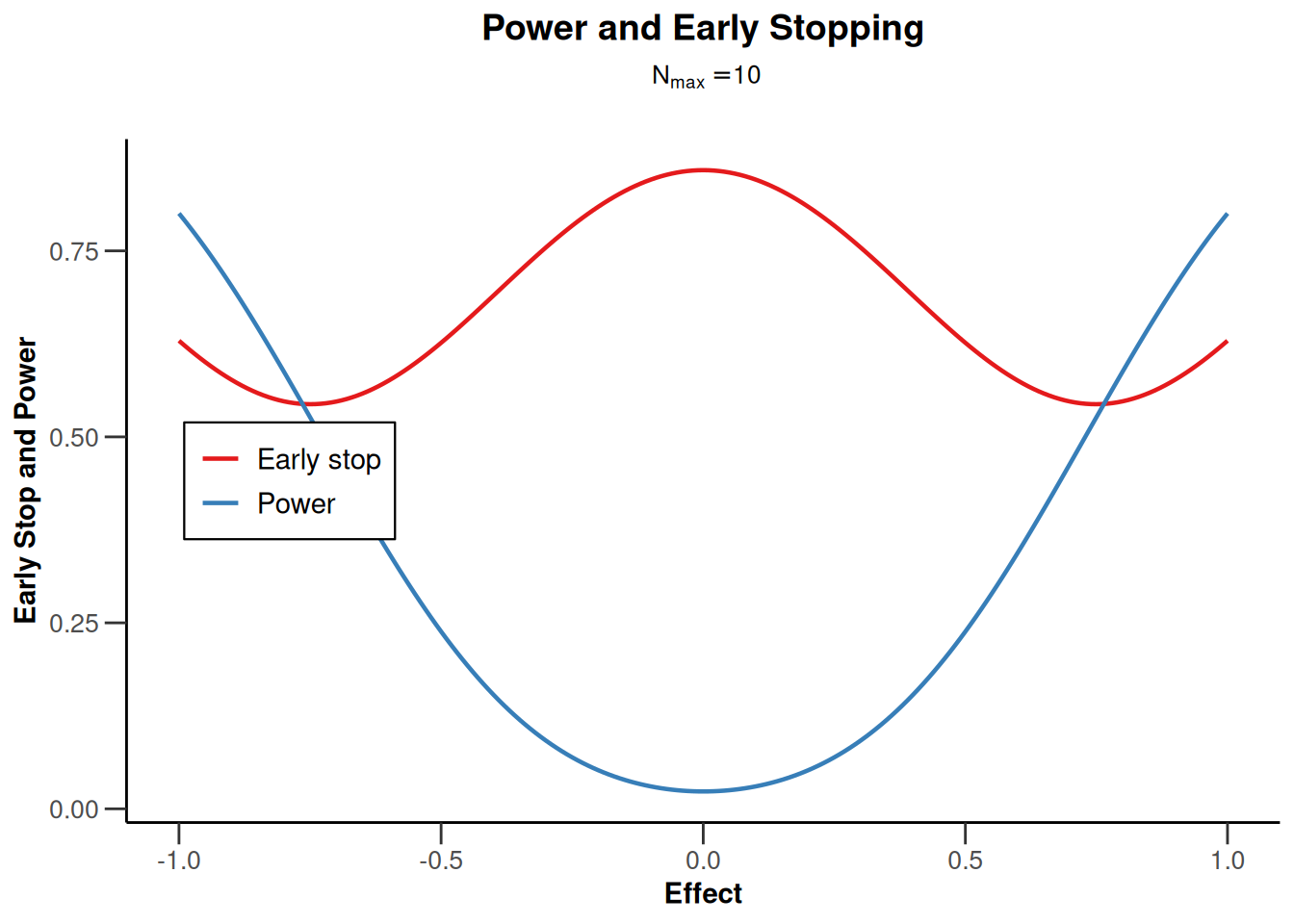
design |> plot(type = 6, nMax = 10)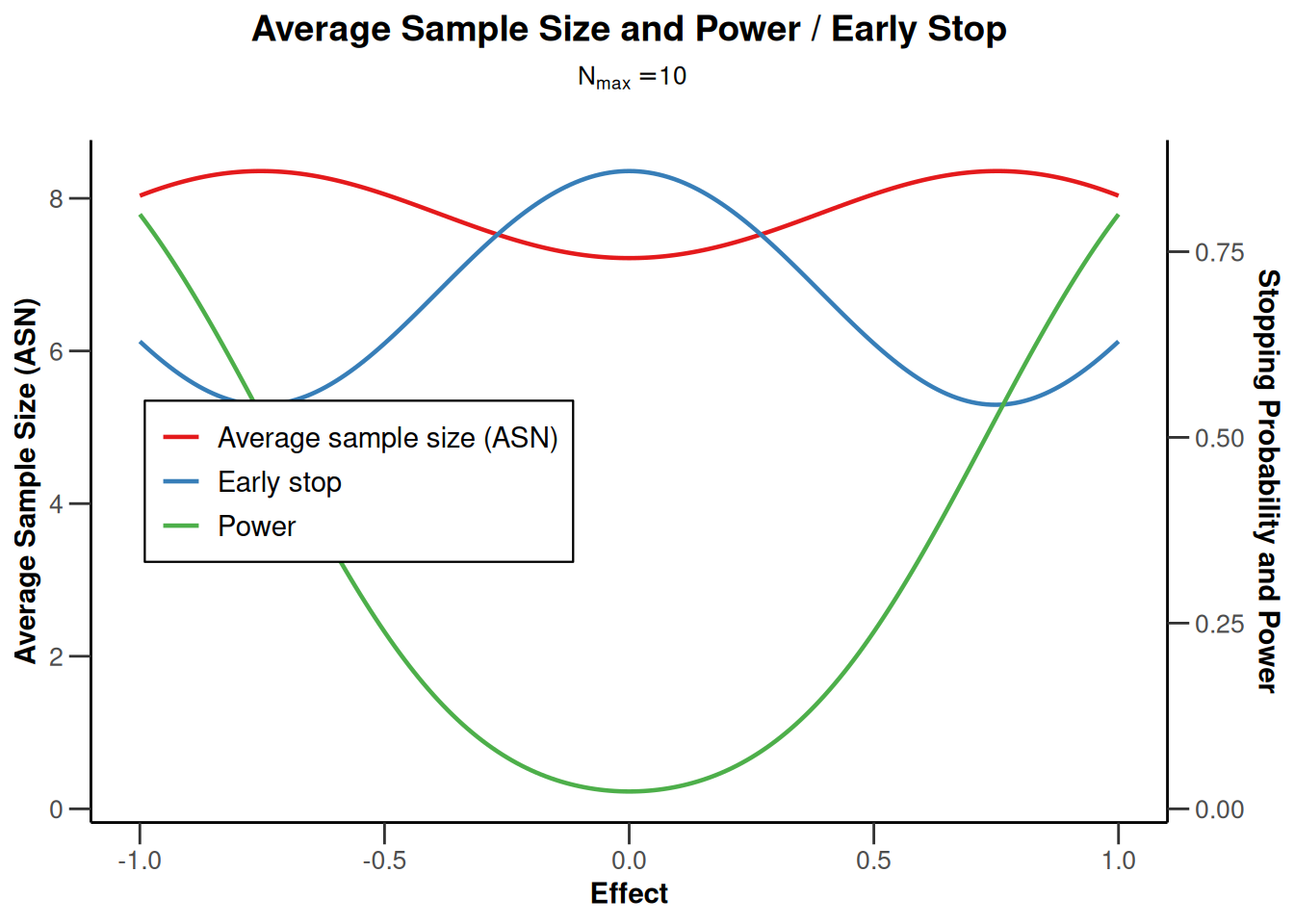
Sample size plots
Sample size means (continuous endpoint)
Sample size means for a one-sided design with futility bounds
sampleSizeMeans1 <- getDesignGroupSequential(
sided = 1,
futilityBounds = c(0, 0.2)) |>
getSampleSizeMeans()
sampleSizeMeans1 |> plot(type = 1)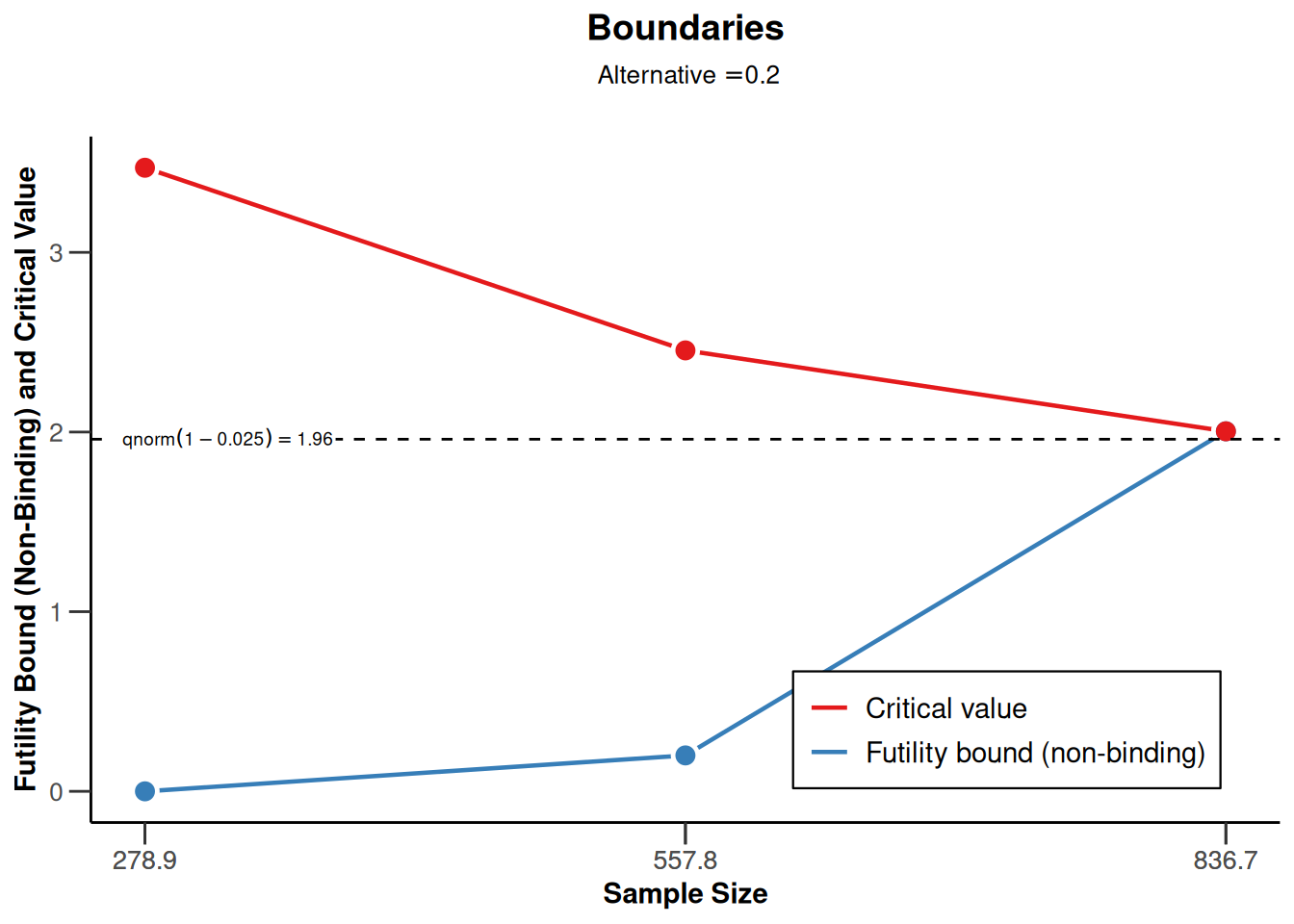
sampleSizeMeans1 |> plot(type = 2)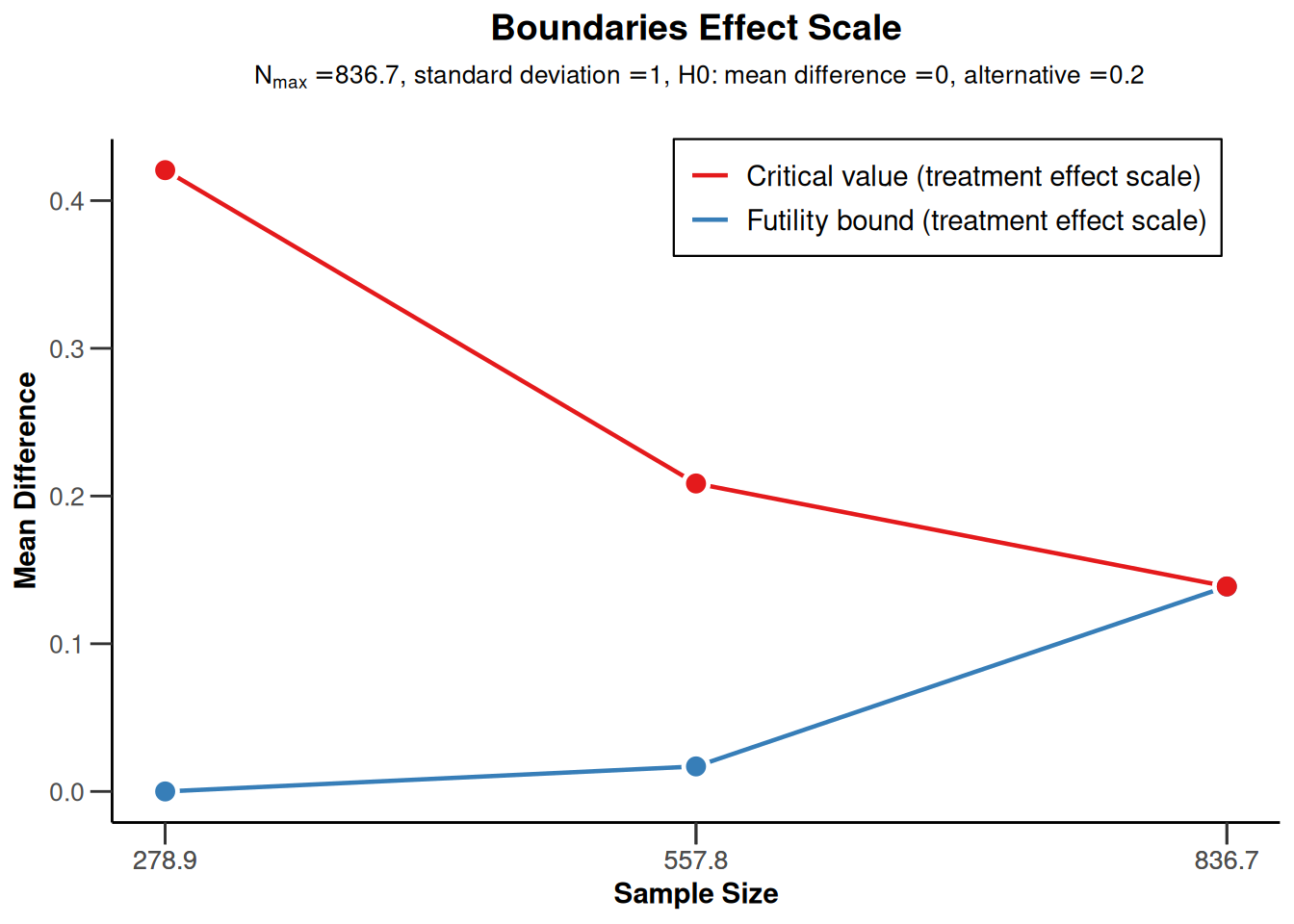
Sample size means for a two-sided design
sampleSizeMeans2 <- getDesignGroupSequential(sided = 2) |>
getSampleSizeMeans()
sampleSizeMeans2 |> plot(type = 1)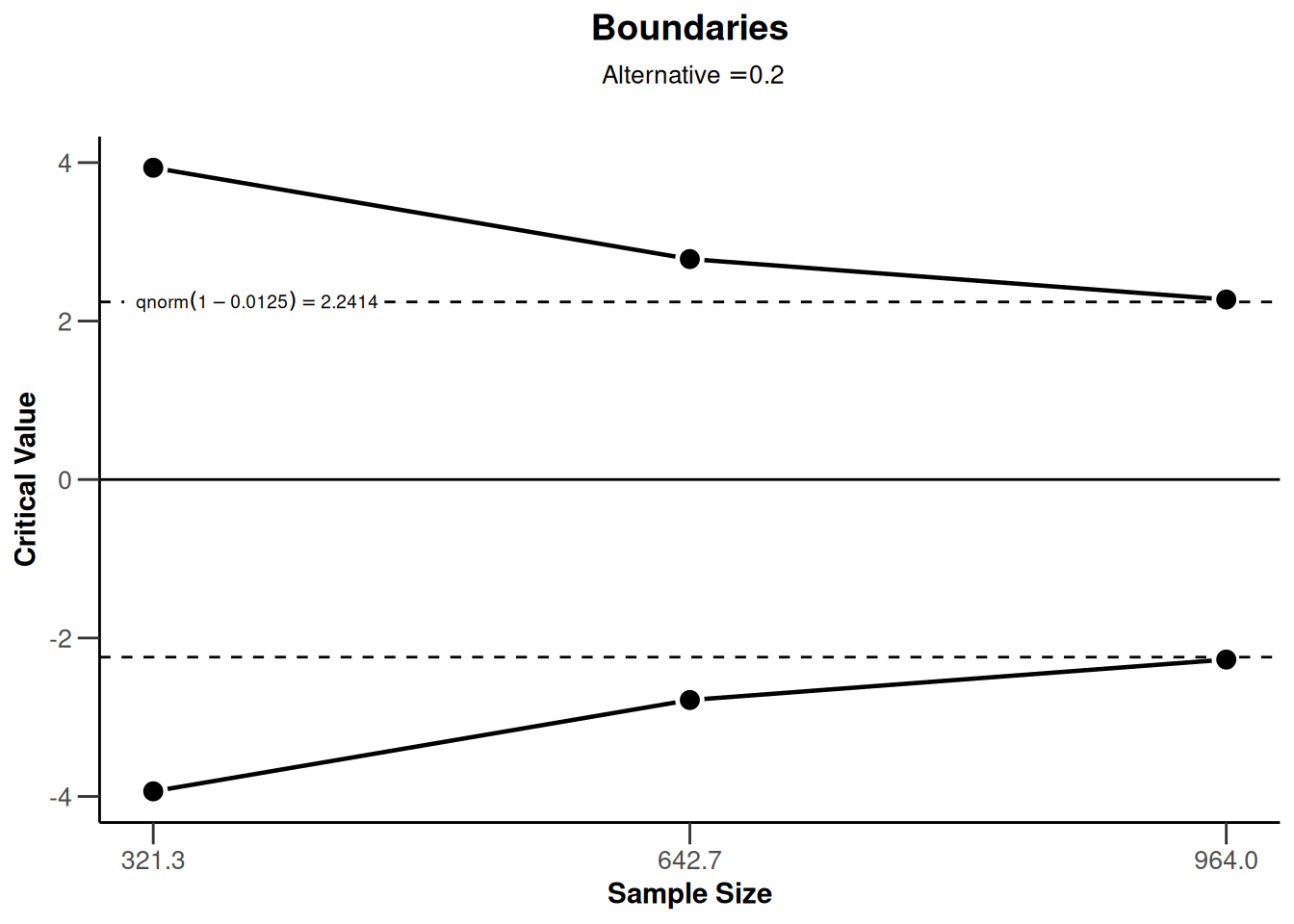
sampleSizeMeans2 |> plot(type = 2)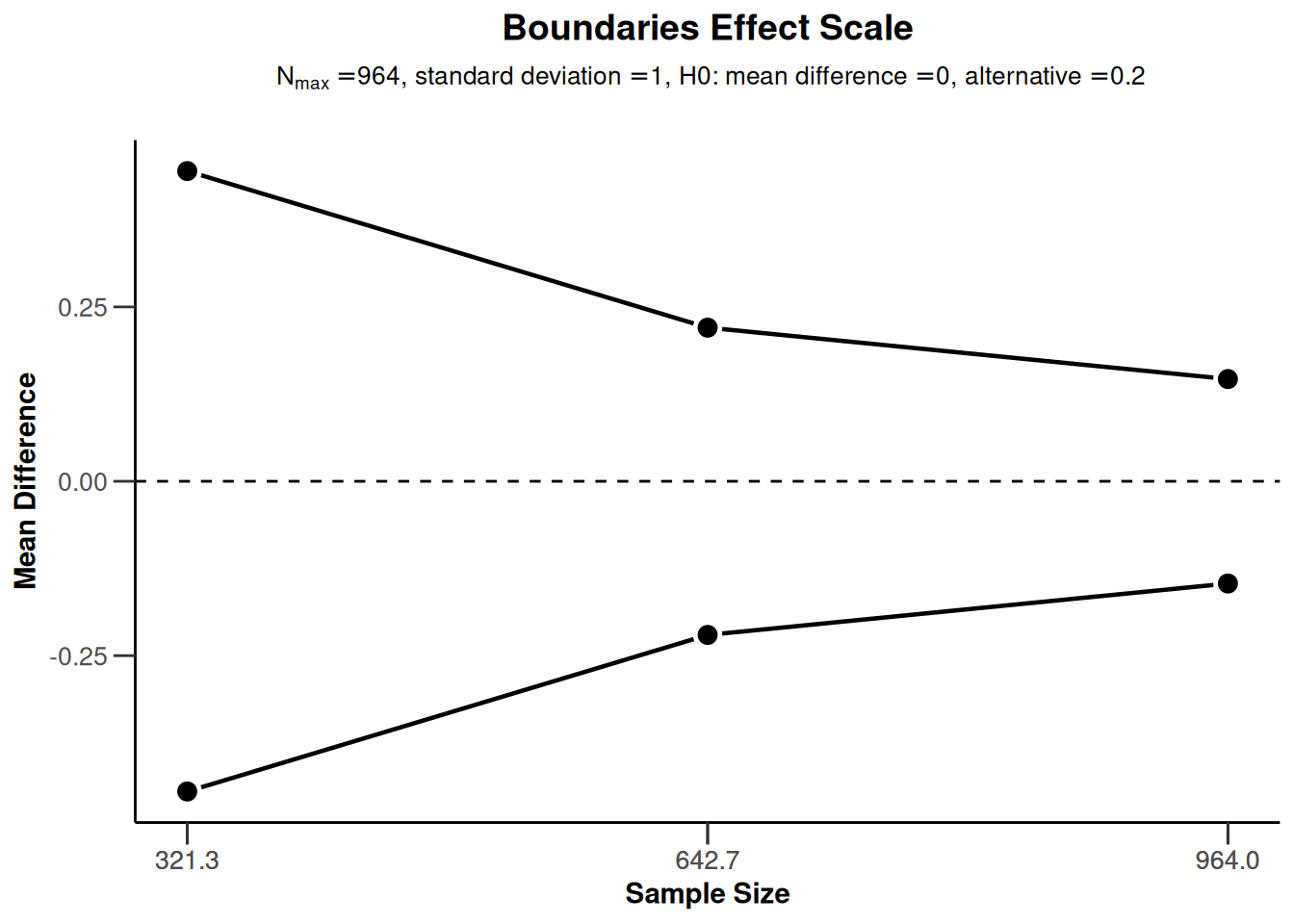
Sample size rates (binary endpoint)
Sample size rates for a one-sided design with futility bounds
sampleSizeRates1 <- getDesignGroupSequential(
sided = 1,
futilityBounds = c(0, 0.1)) |>
getSampleSizeRates()
sampleSizeRates1 |> plot(type = 1)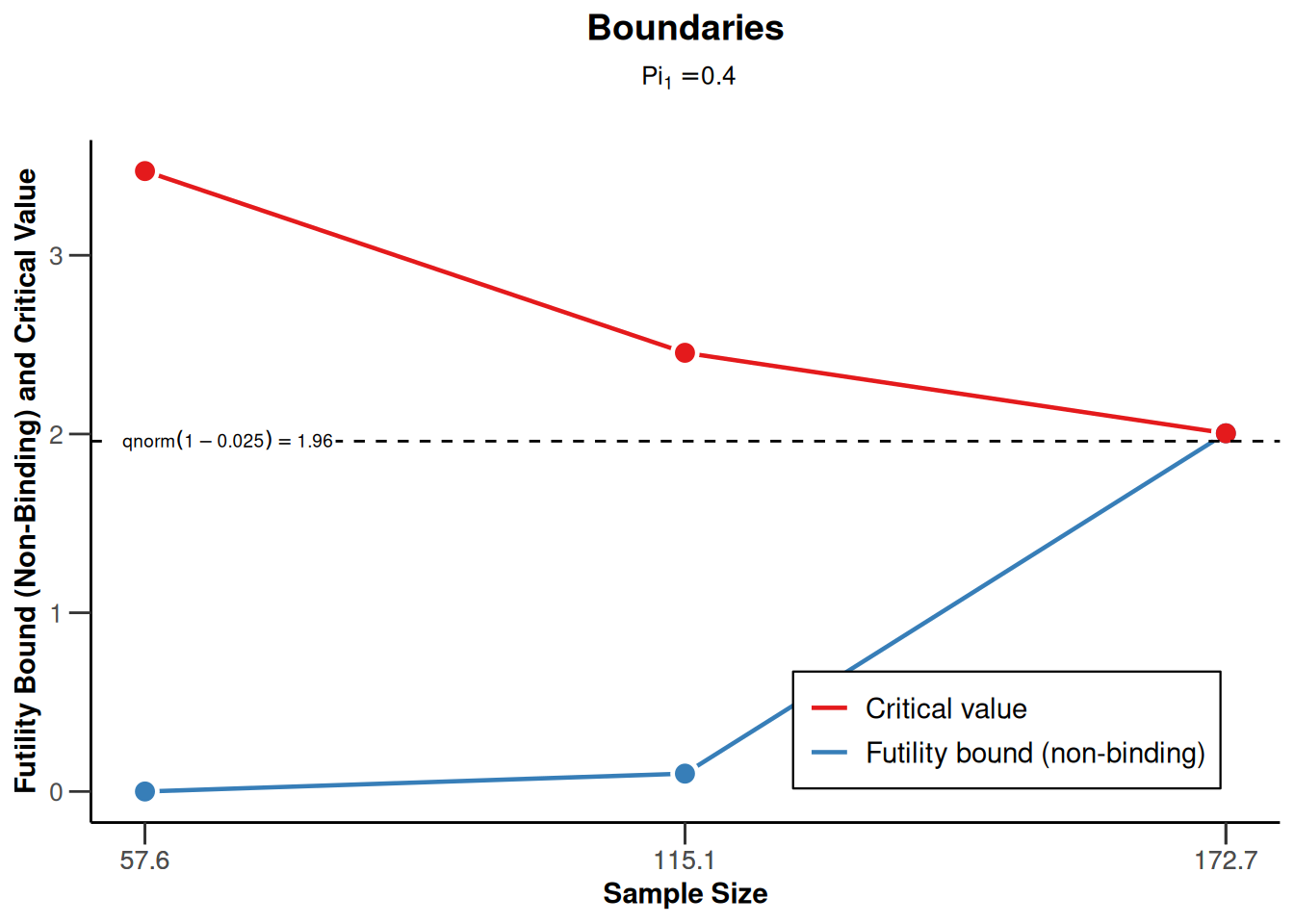
sampleSizeRates1 |> plot(type = 2)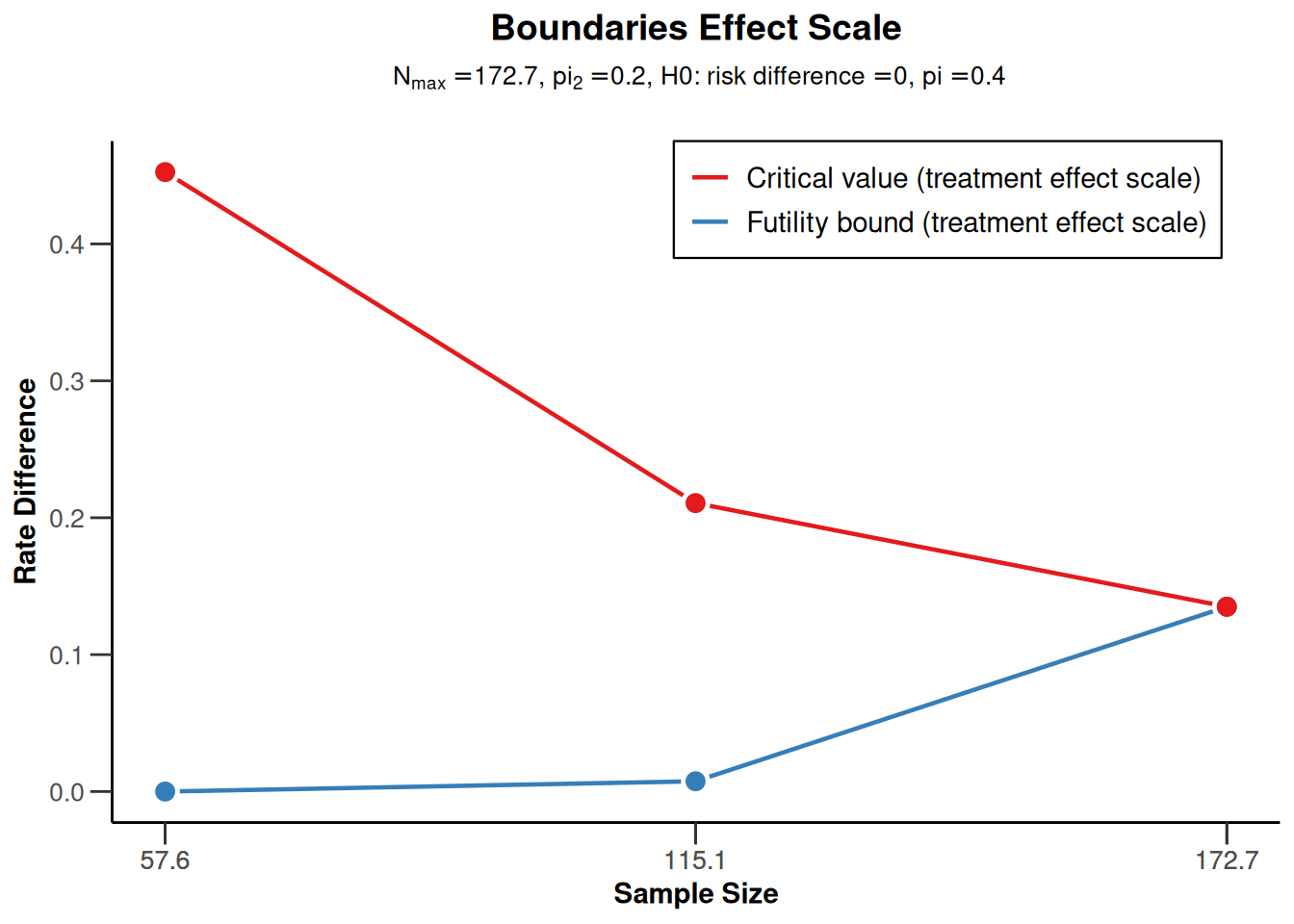
Sample size rates for a two-sided design
sampleSizeRates2 <- getDesignGroupSequential(sided = 2) |>
getSampleSizeRates()
sampleSizeRates2 |> plot(type = 1)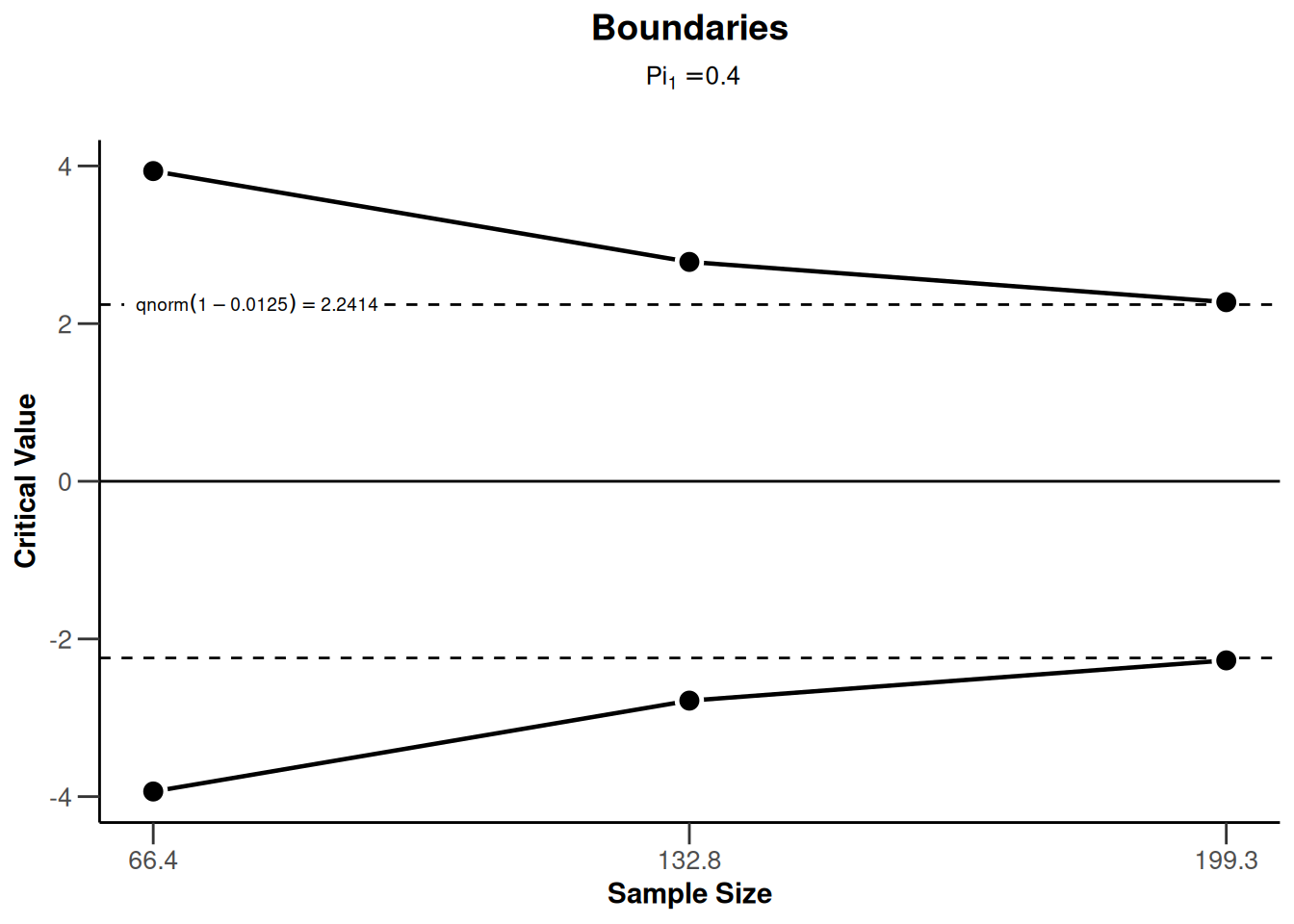
sampleSizeRates2 |> plot(type = 2)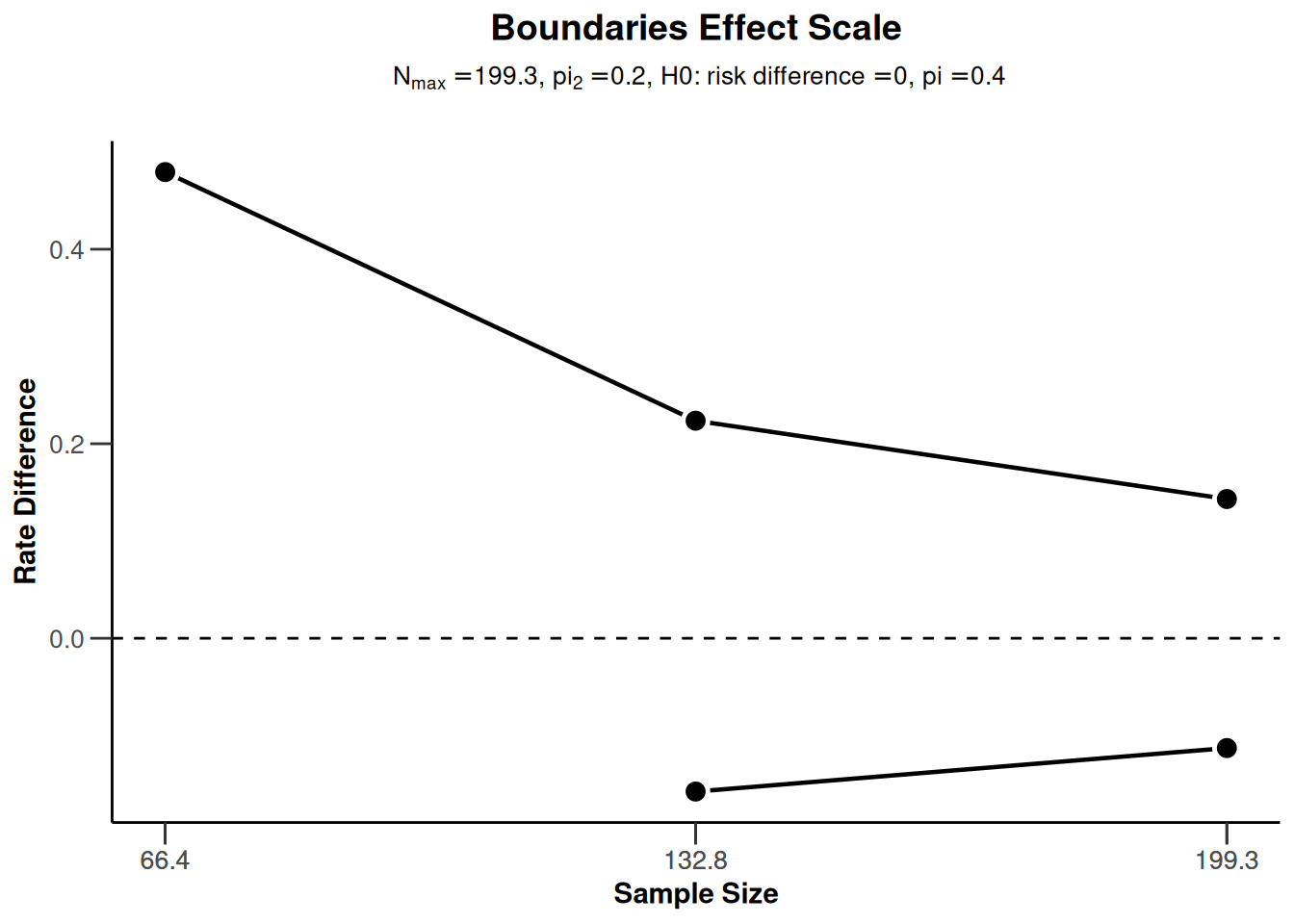
Sample size survival (survival endpoint)
Sample size survival for a one-sided design with futility bounds
design <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(-1, 0.5)
)
piecewiseSurvivalTime <- list(
"0 - <6" = 0.025,
"6 - <9" = 0.04,
"9 - <15" = 0.015,
"15 - <21" = 0.01,
">= 21" = 0.007
)
sampleSizeSurvival1 <- getSampleSizeSurvival(
design = design,
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
piecewiseSurvivalTime = piecewiseSurvivalTime,
hazardRatio = c(0.5, 0.9)
)
sampleSizeSurvival1 |> plot(type = 1)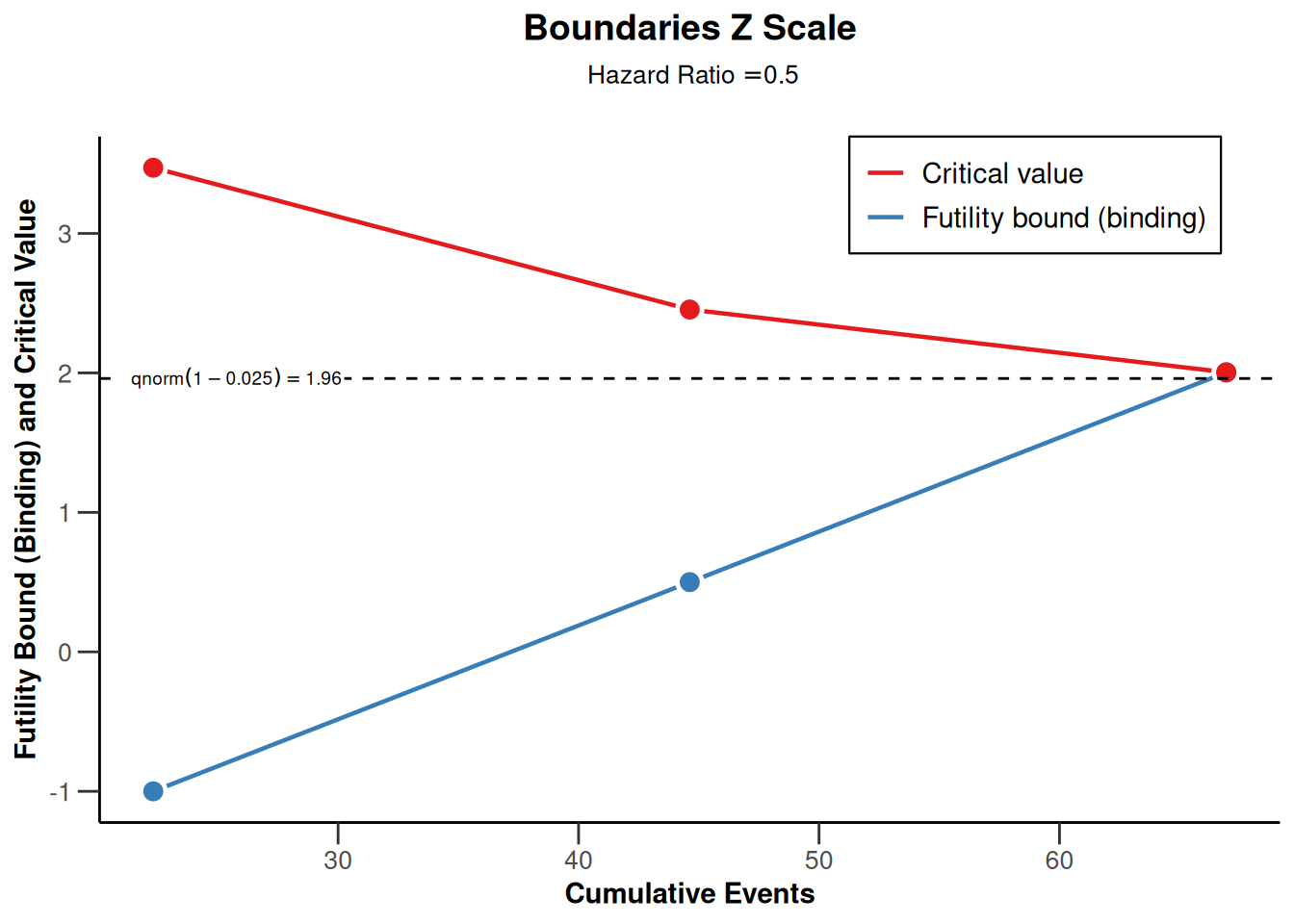
sampleSizeSurvival1 |> plot(type = 2)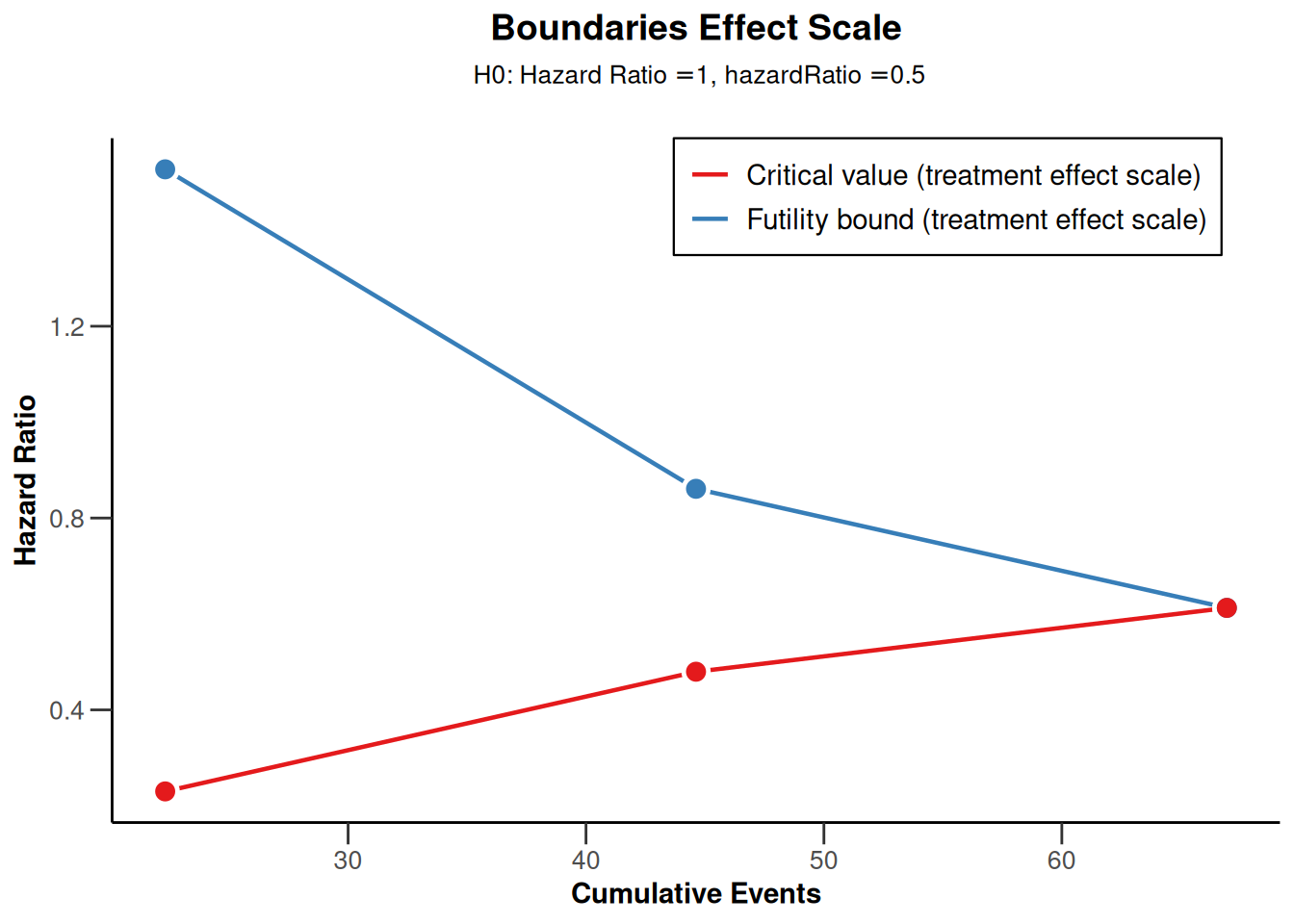
sampleSizeSurvival1 |> plot(type = 13, legendPosition = 1)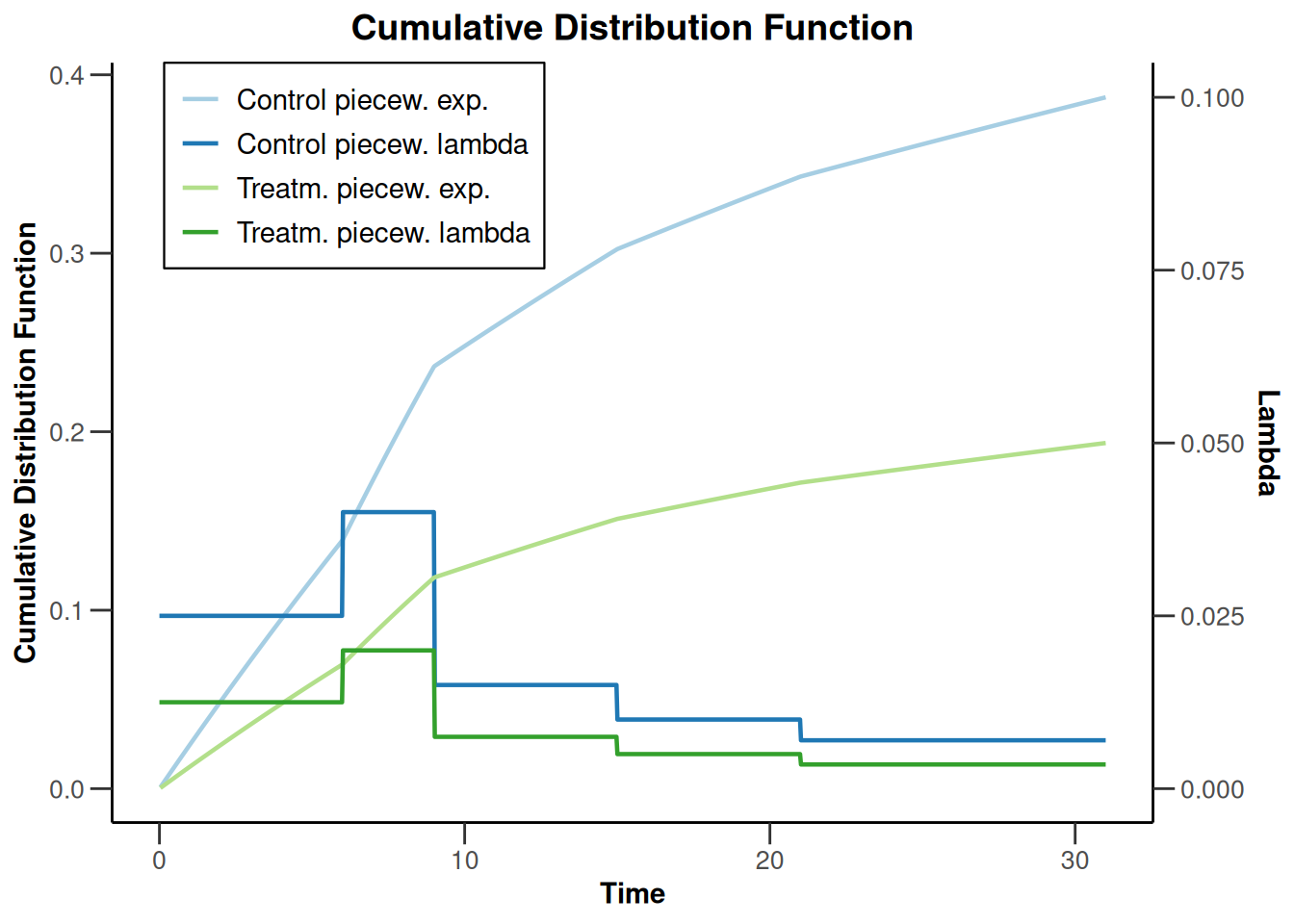
sampleSizeSurvival1 |> plot(type = 14)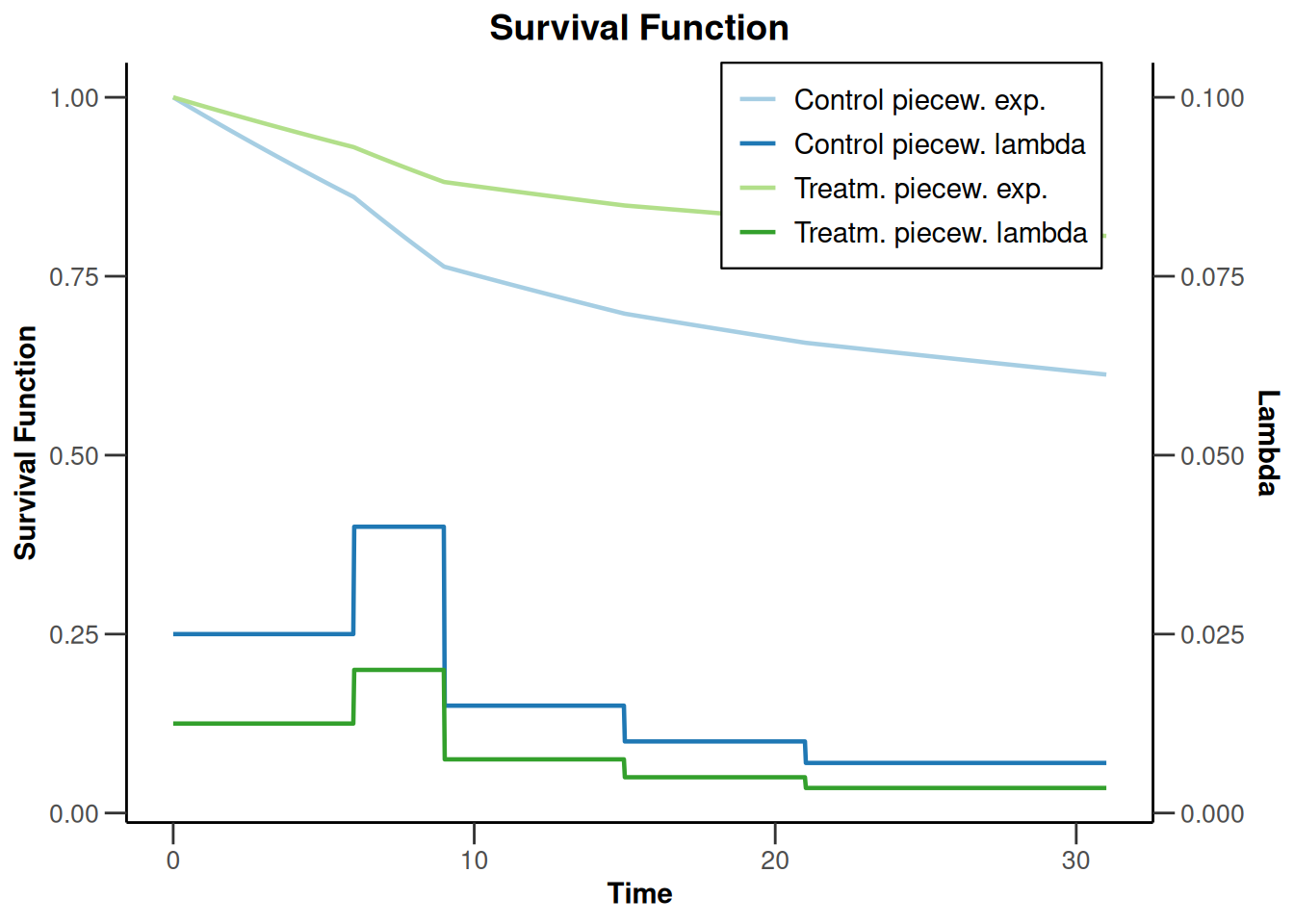
Sample size survival for a two-sided design
design <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 2,
twoSidedPower = TRUE
)
piecewiseSurvivalTime <- list(
"0 - <14" = 0.015,
"14 - <24" = 0.01,
"24 - <44" = 0.005,
">= 44" = 0.0025
)
sampleSizeSurvival2 <- getSampleSizeSurvival(
design = design,
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
piecewiseSurvivalTime = piecewiseSurvivalTime,
maxNumberOfSubjects = 0,
hazardRatio = c(0.1, 0.2, 0.5, 0.6)
)
sampleSizeSurvival2 |> plot(type = 1)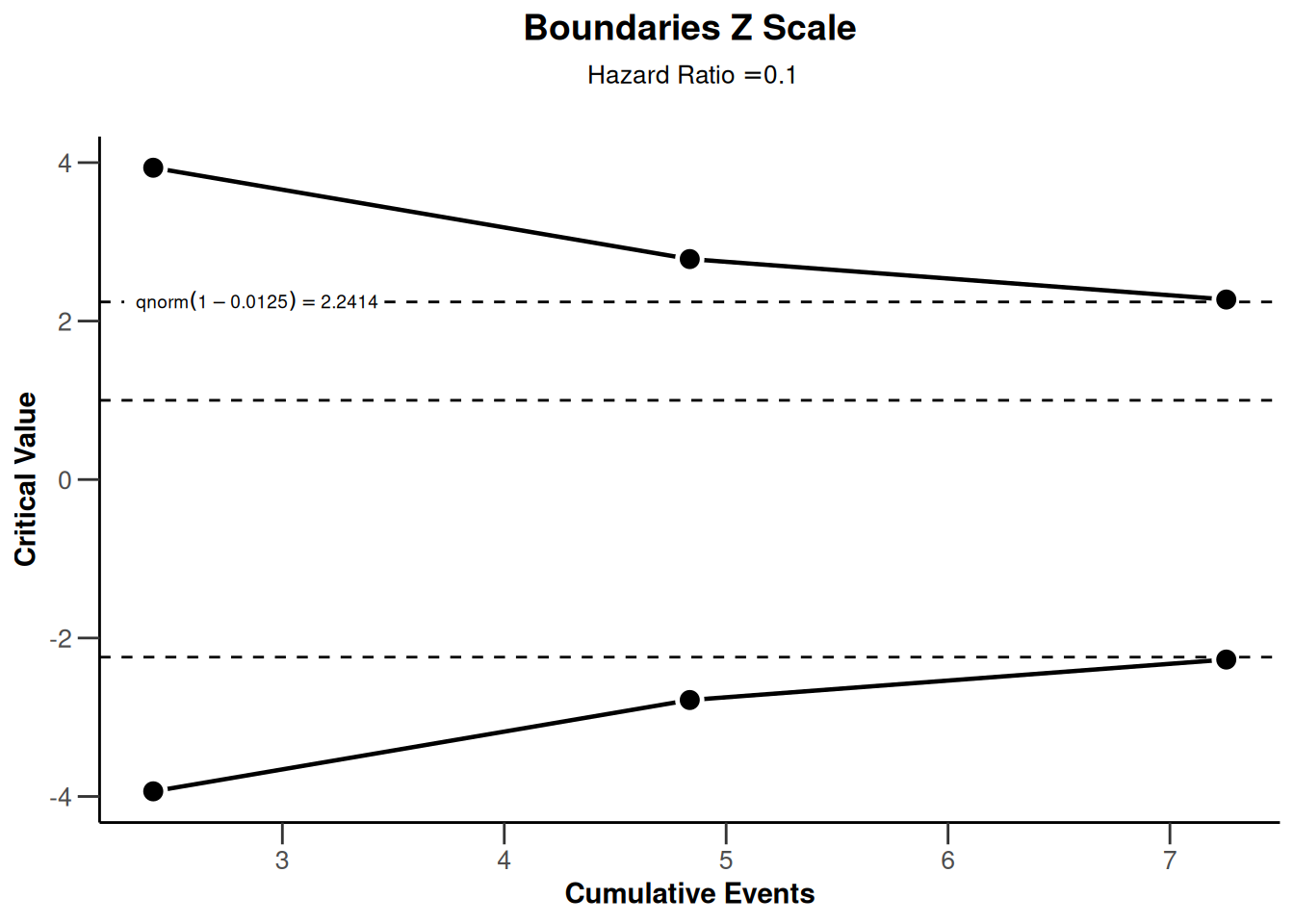
sampleSizeSurvival2 |> plot(type = 2)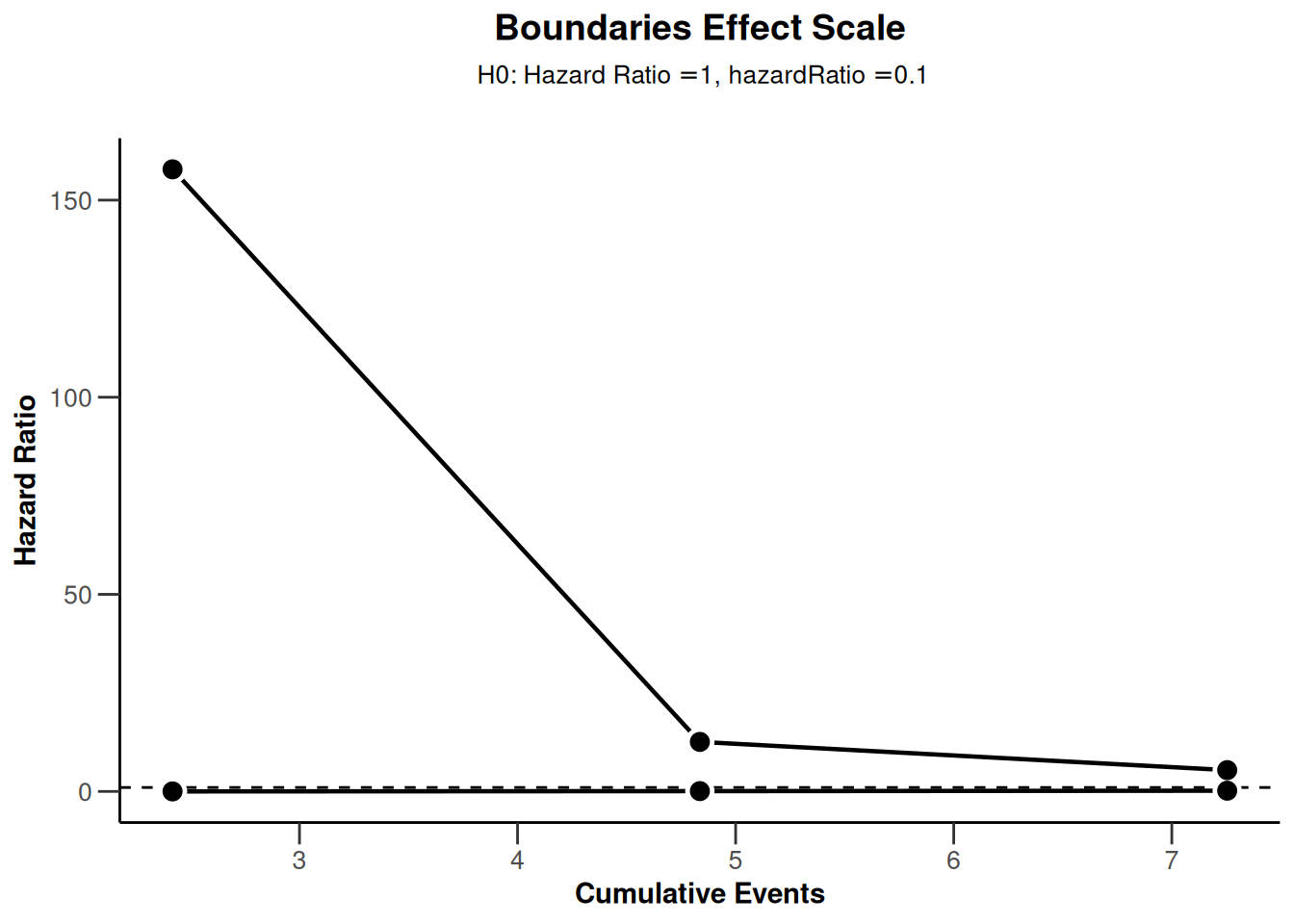
sampleSizeSurvival2 |> plot(type = 13, legendPosition = 1)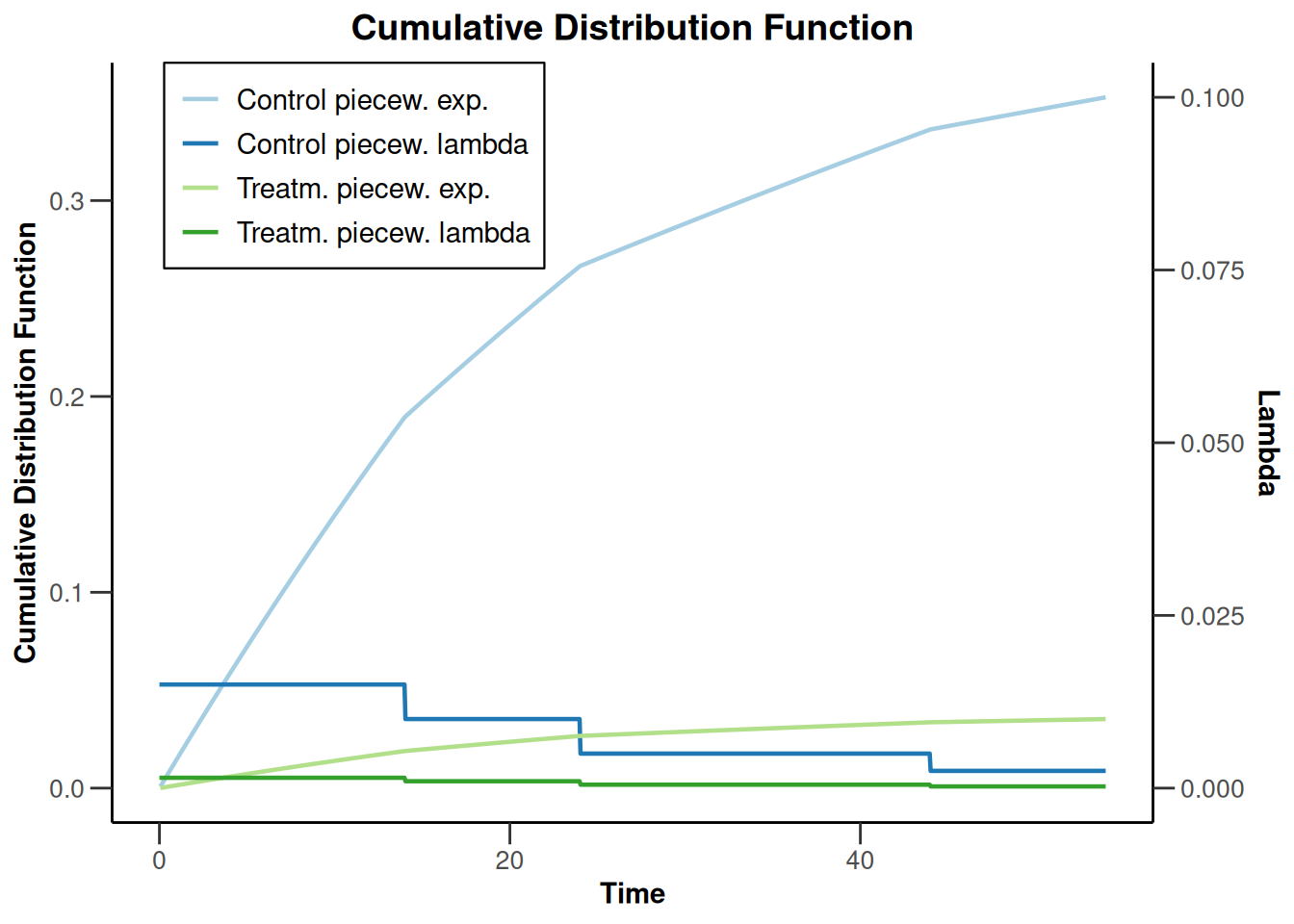
sampleSizeSurvival2 |> plot(type = 14)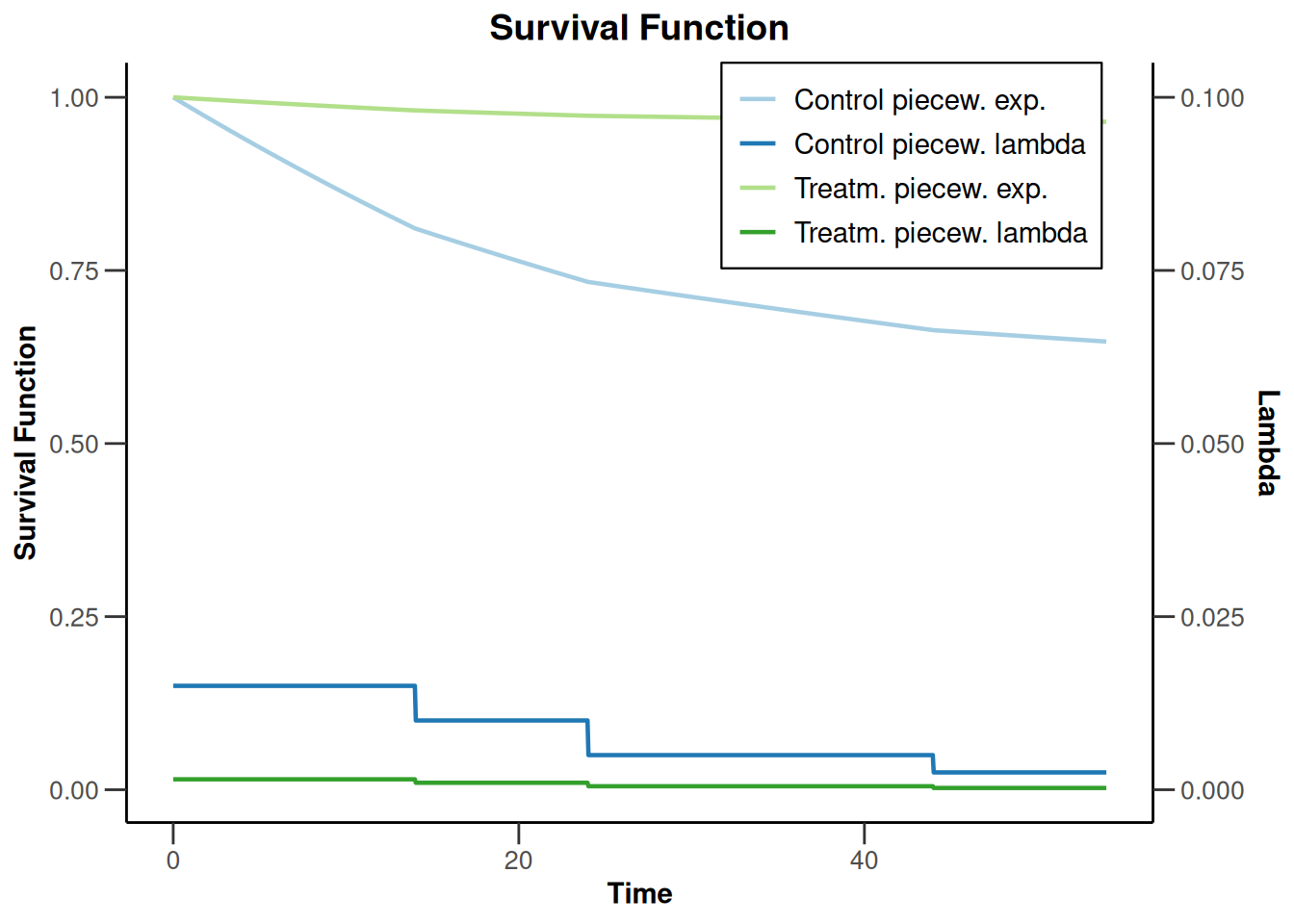
Power plots
Power means (continuous endpoint)
Power means for a one-sided design with futility bounds
powerMeans1 <- getDesignGroupSequential(
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(0, 0.5)) |>
getPowerMeans(
groups = 1,
meanRatio = FALSE,
thetaH0 = 0,
alternative = c(-1, 4),
stDev = 2,
normalApproximation = FALSE,
maxNumberOfSubjects = 40
)
powerMeans1 |> plot(type = 1)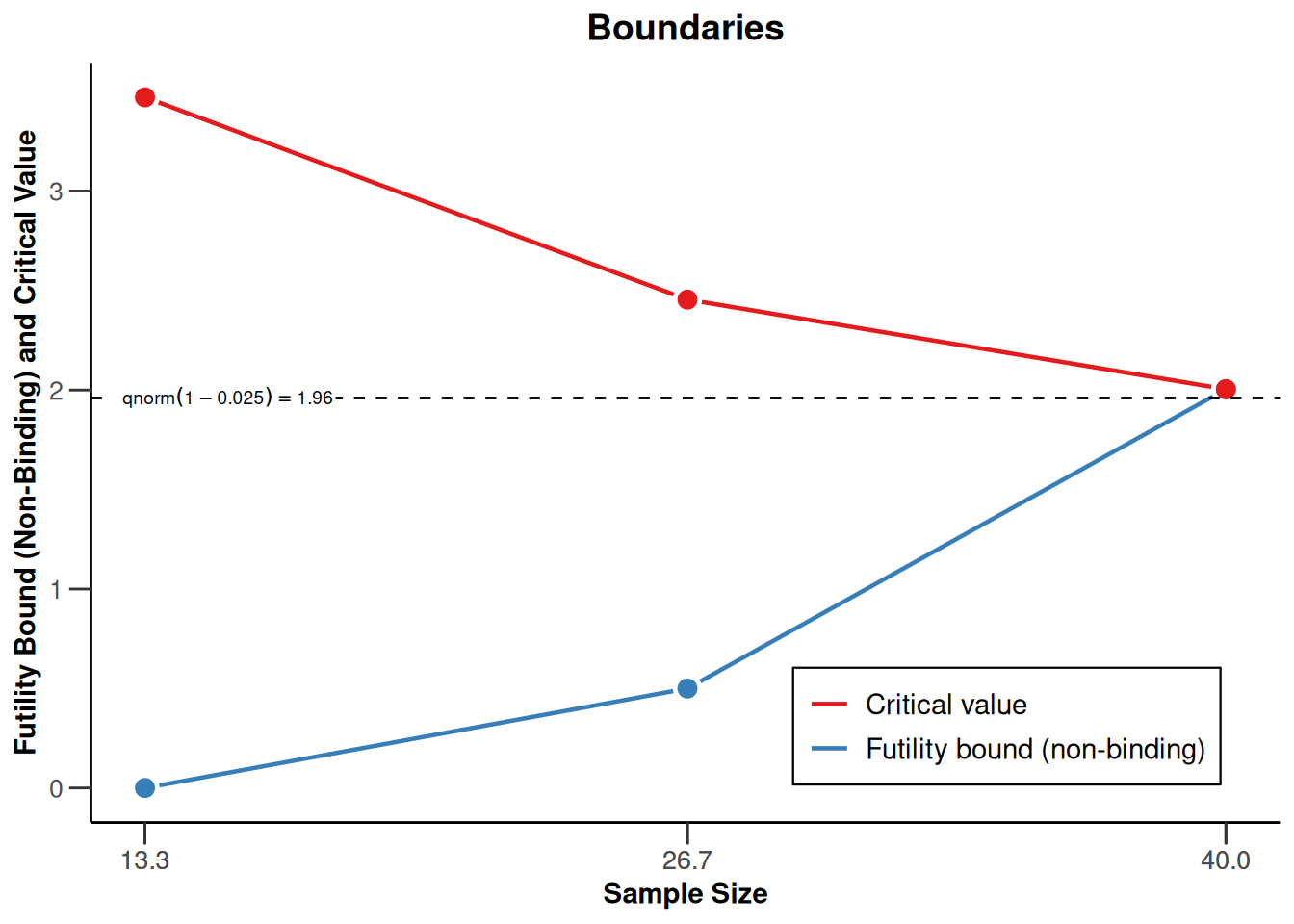
powerMeans1 |> plot(type = 2)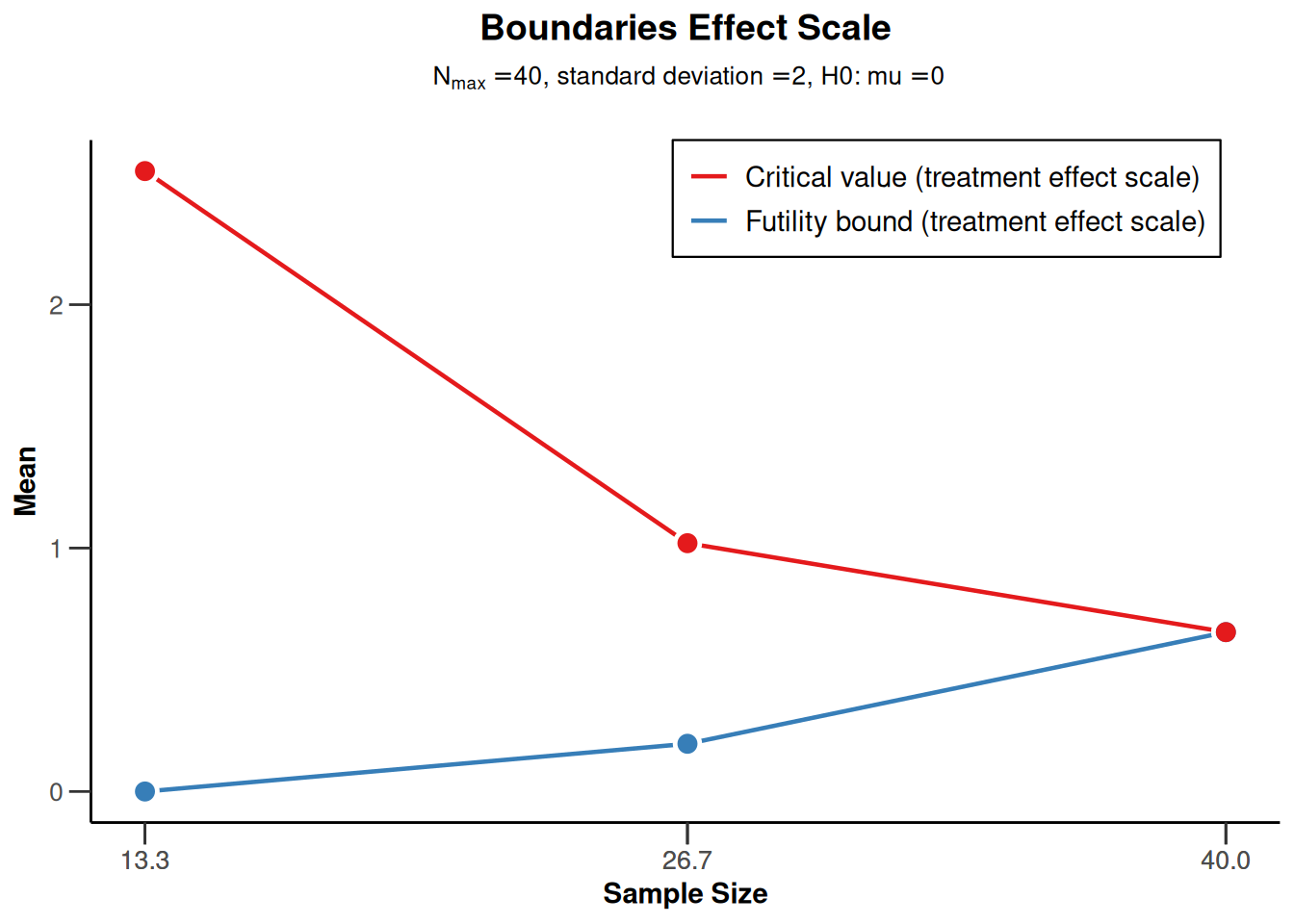
powerMeans1 |> plot(type = 5)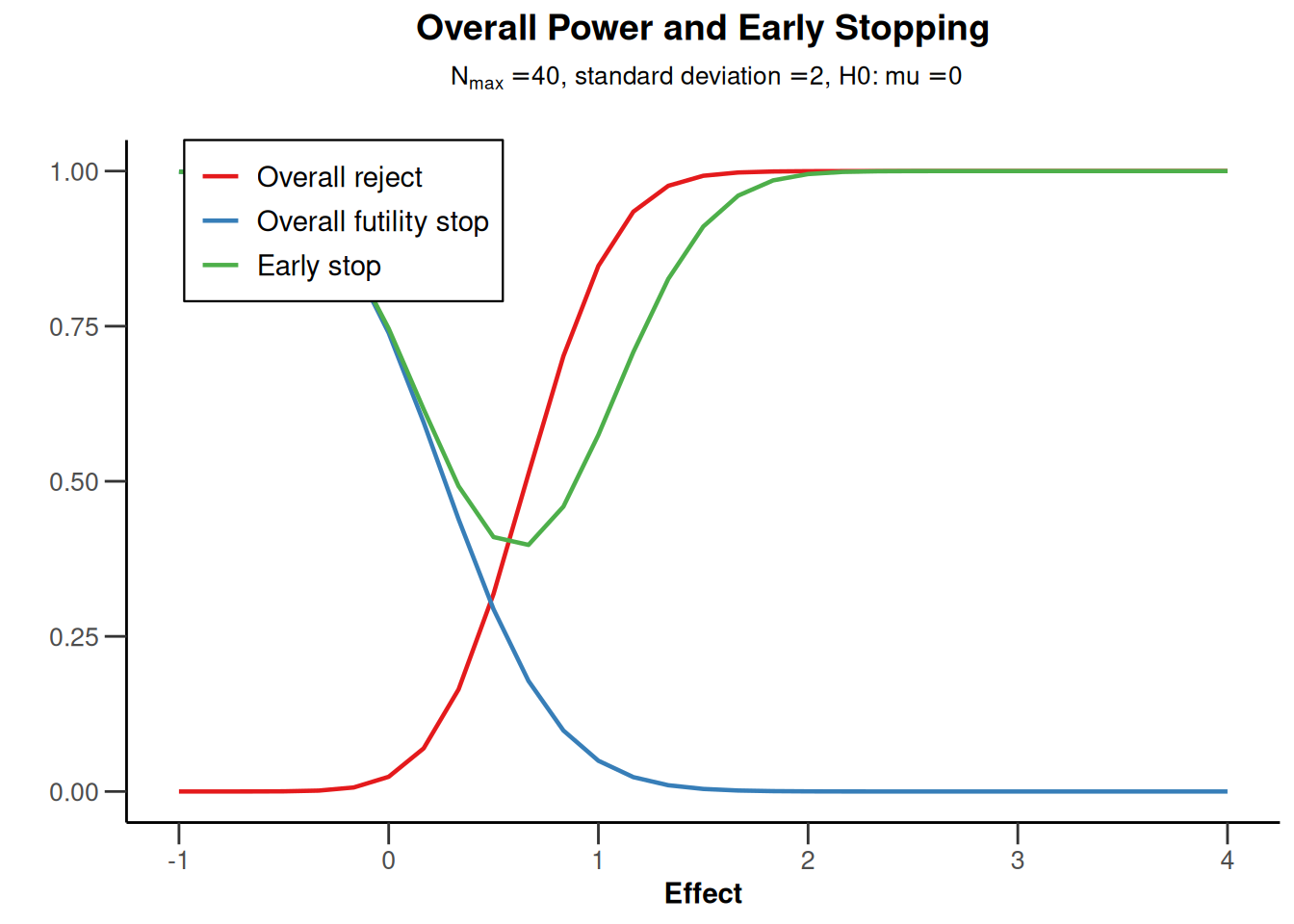
powerMeans1 |> plot(type = 6)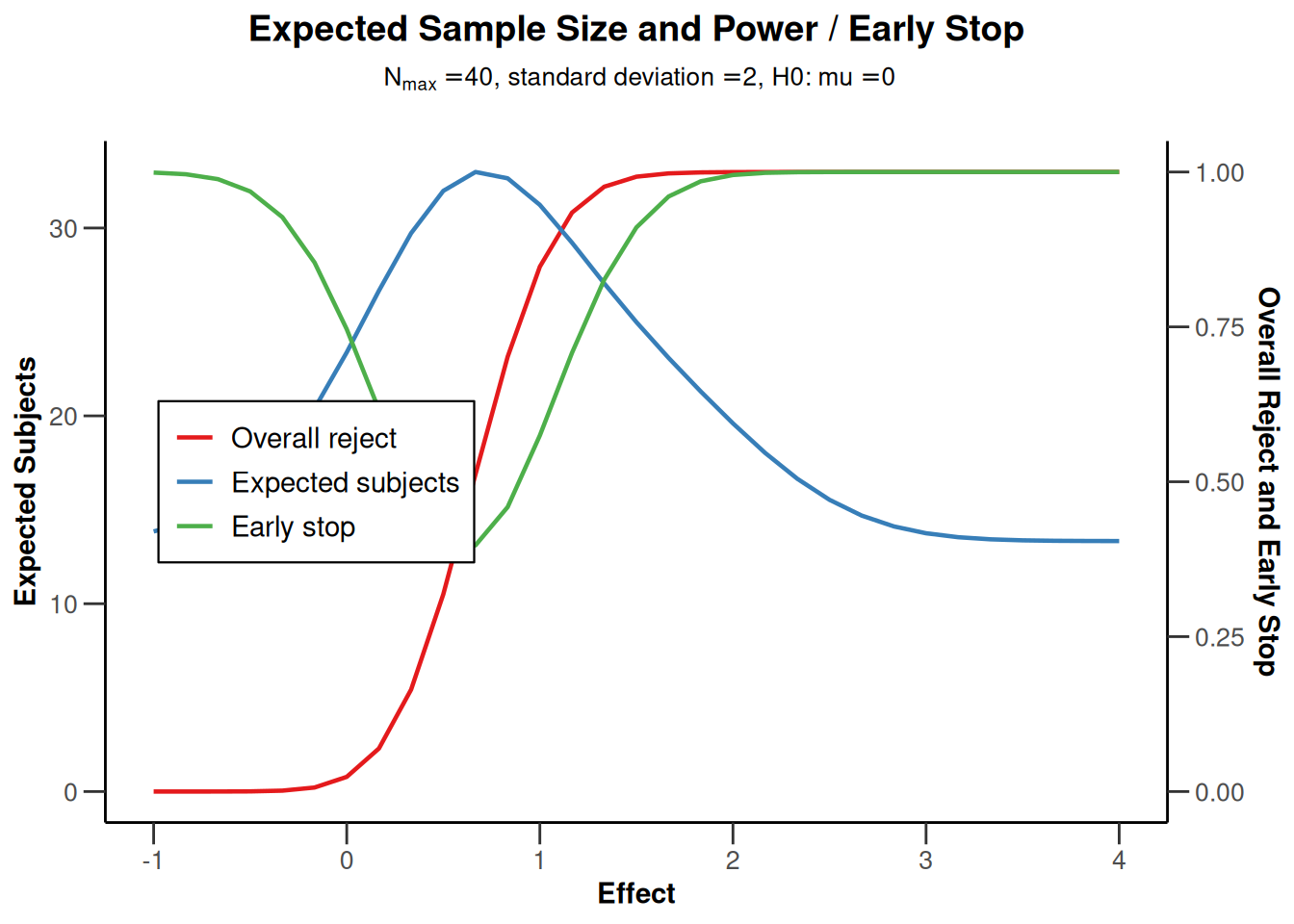
Power means for a two-sided design
powerMeans2 <- getDesignGroupSequential(
typeOfDesign = "OF",
sided = 2,
twoSidedPower = TRUE) |>
getPowerMeans(maxNumberOfSubjects = 30)
powerMeans2 |> plot(type = 1)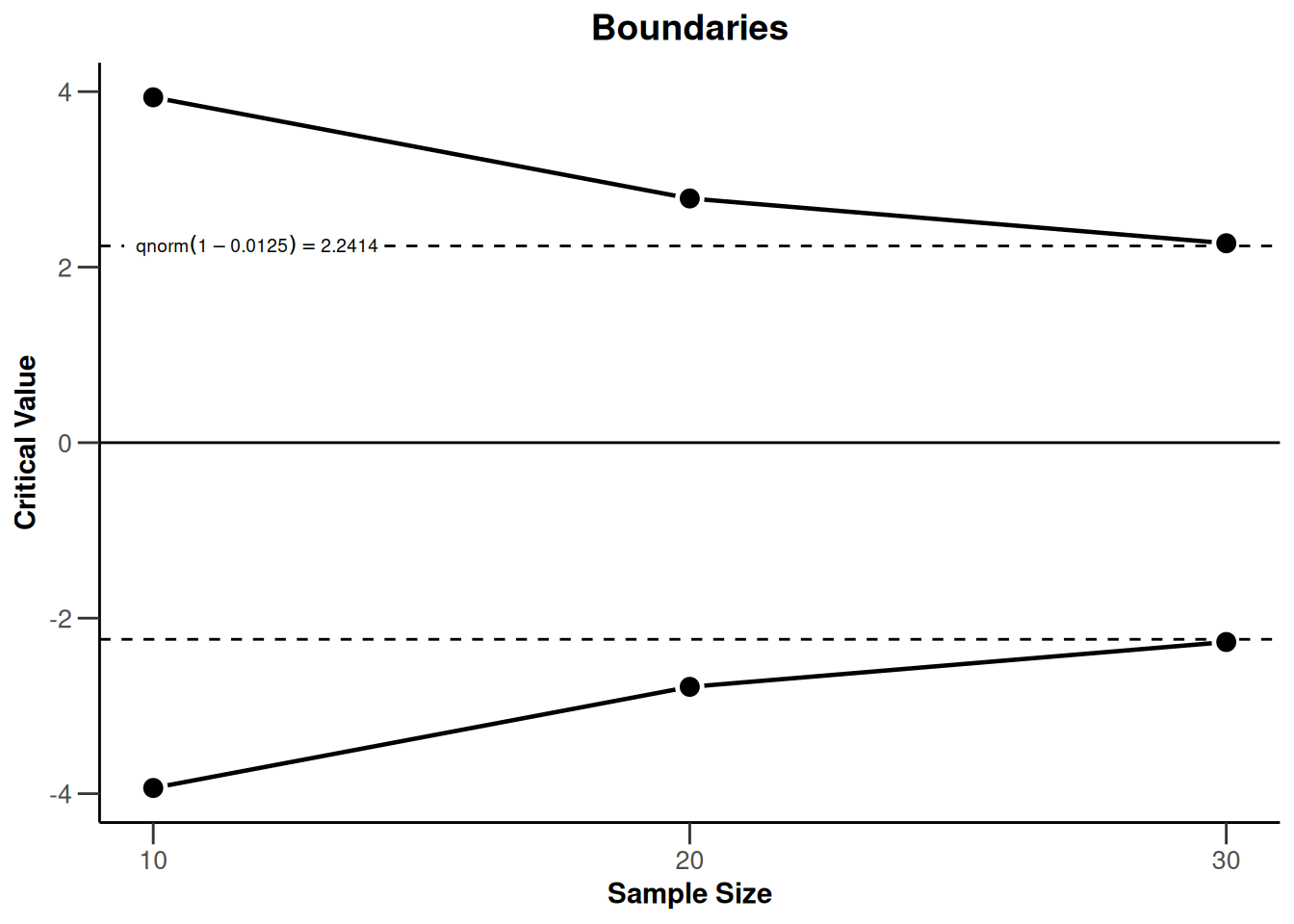
powerMeans2 |> plot(type = 2)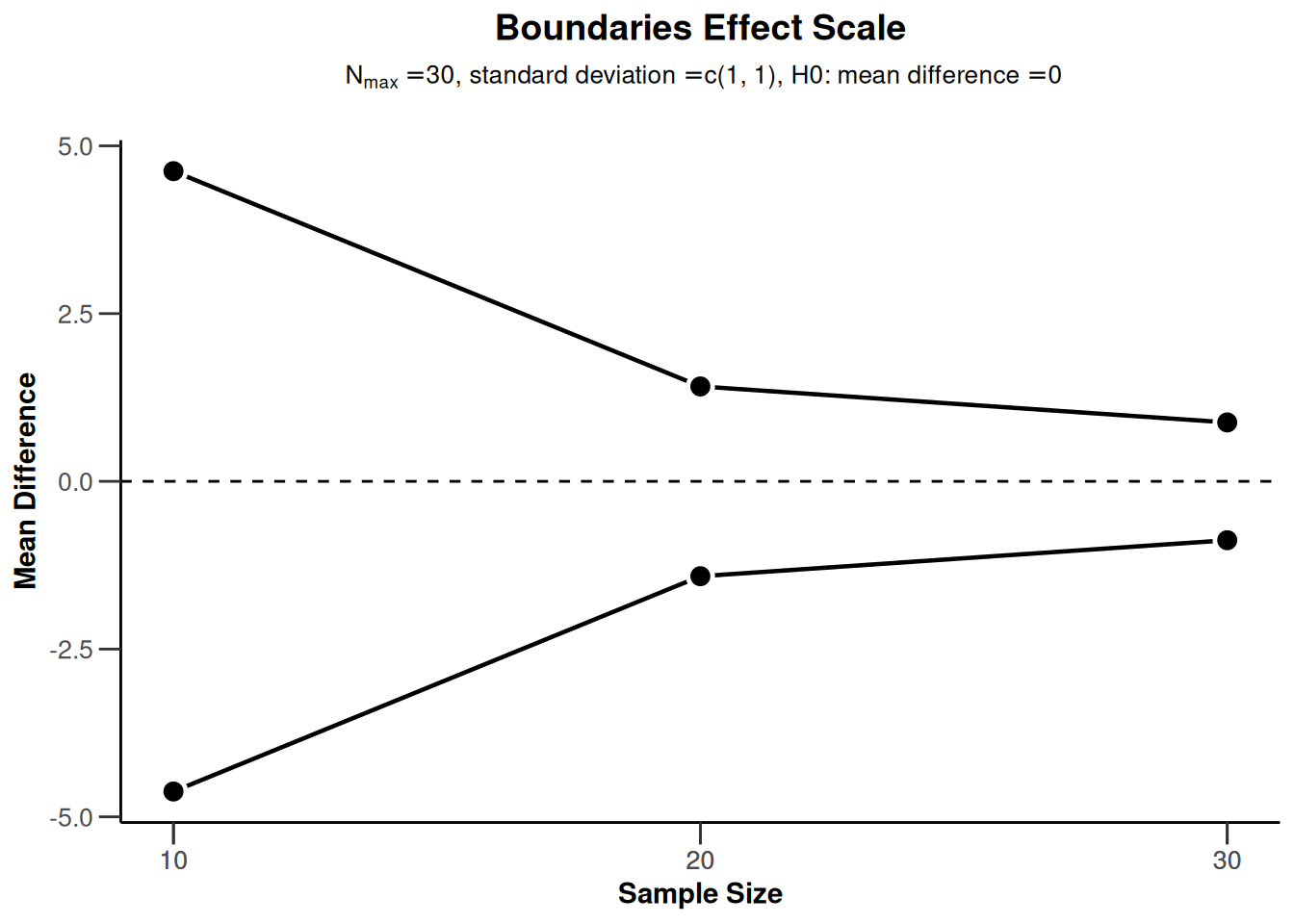
powerMeans2 |> plot(type = 5)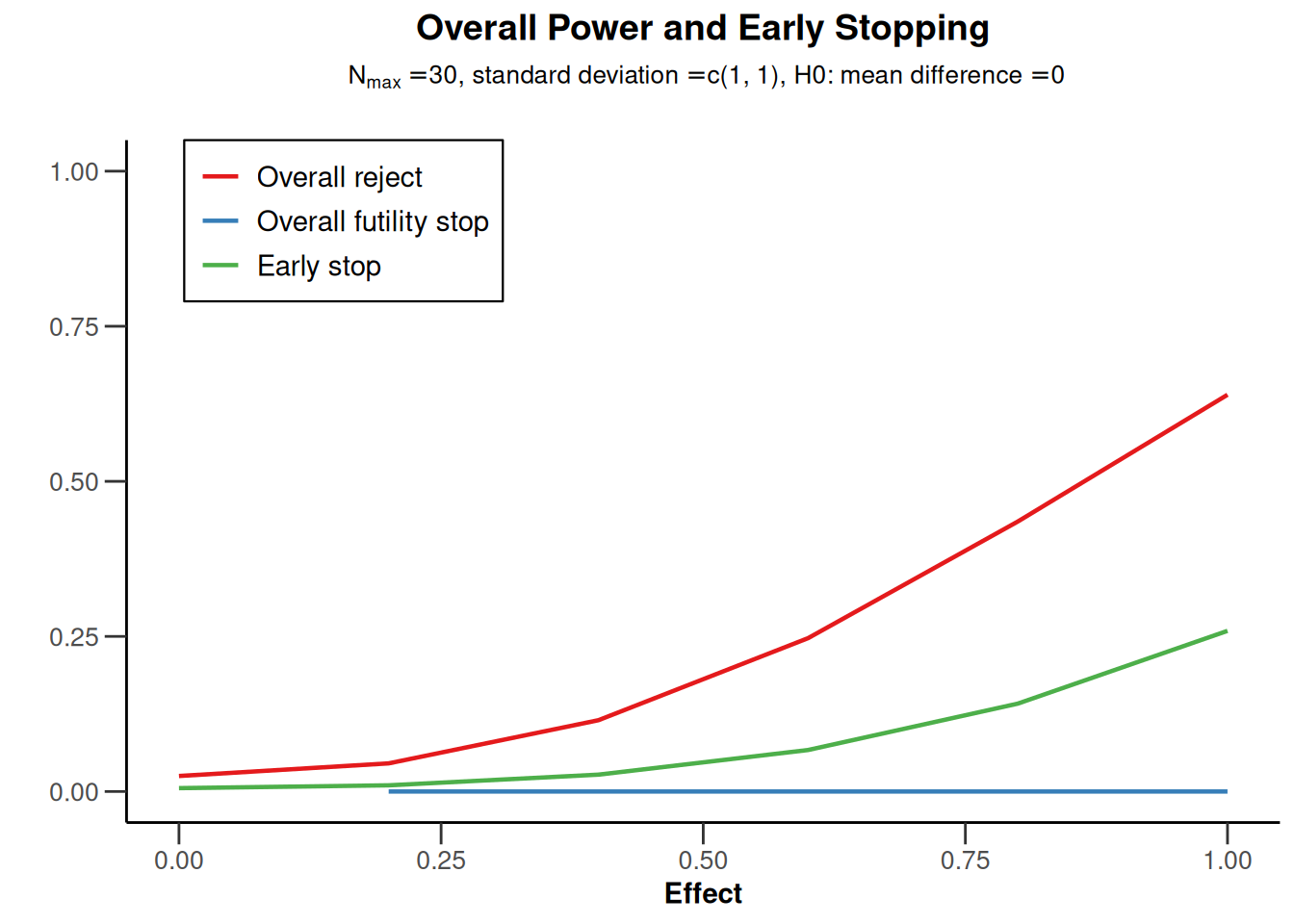
powerMeans2 |> plot(type = 6)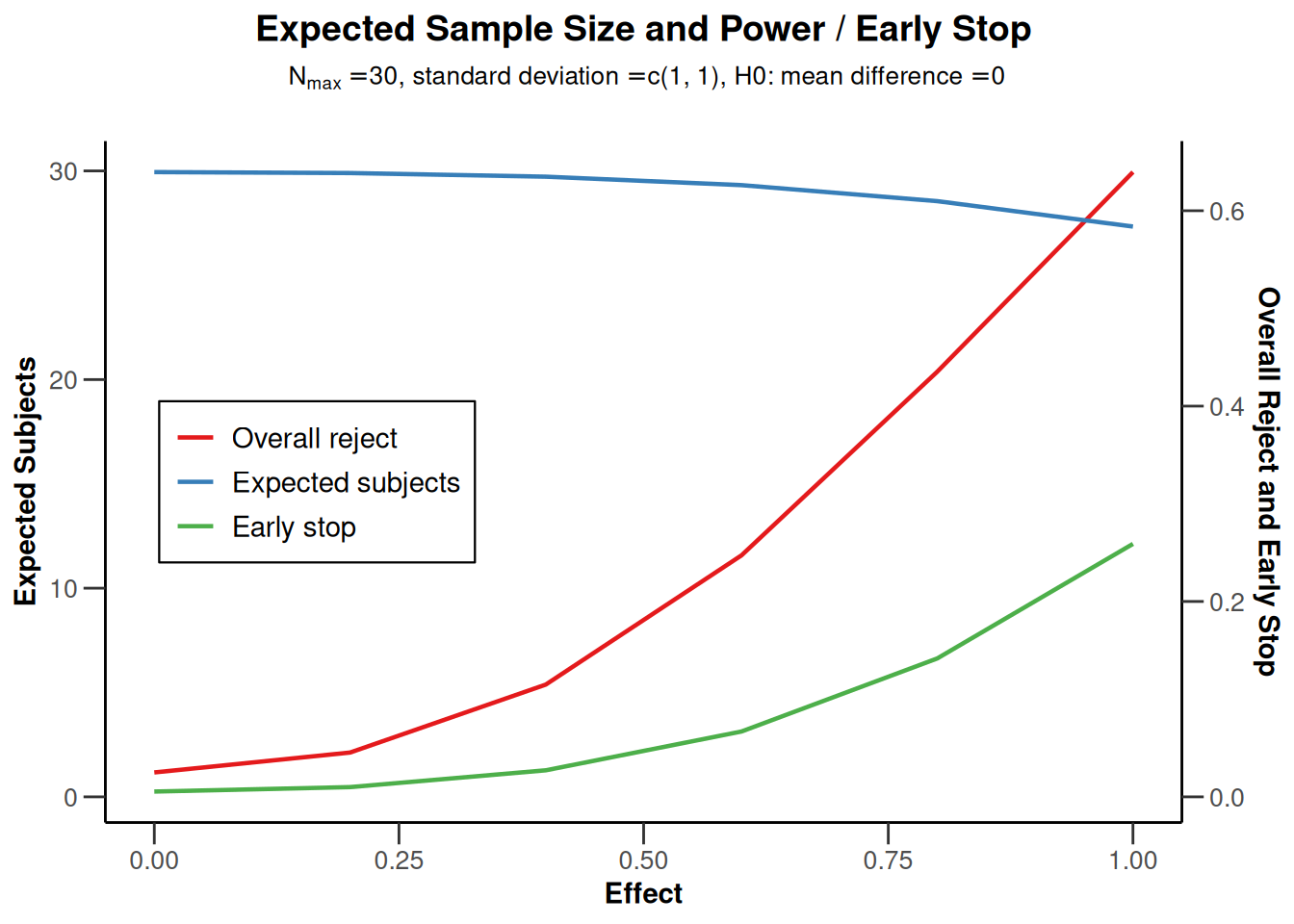
Power rates (binary endpoint)
Power rates for a one-sided design with futility bounds
powerRates1 <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(-1, 0.5)) |>
getPowerRates(
groups = 2,
riskRatio = TRUE,
thetaH0 = 0.2,
allocationRatioPlanned = 1,
pi1 = c(0.1, 0.4),
pi2 = 0.2,
maxNumberOfSubjects = 80
)
powerRates1 |> plot(type = 1)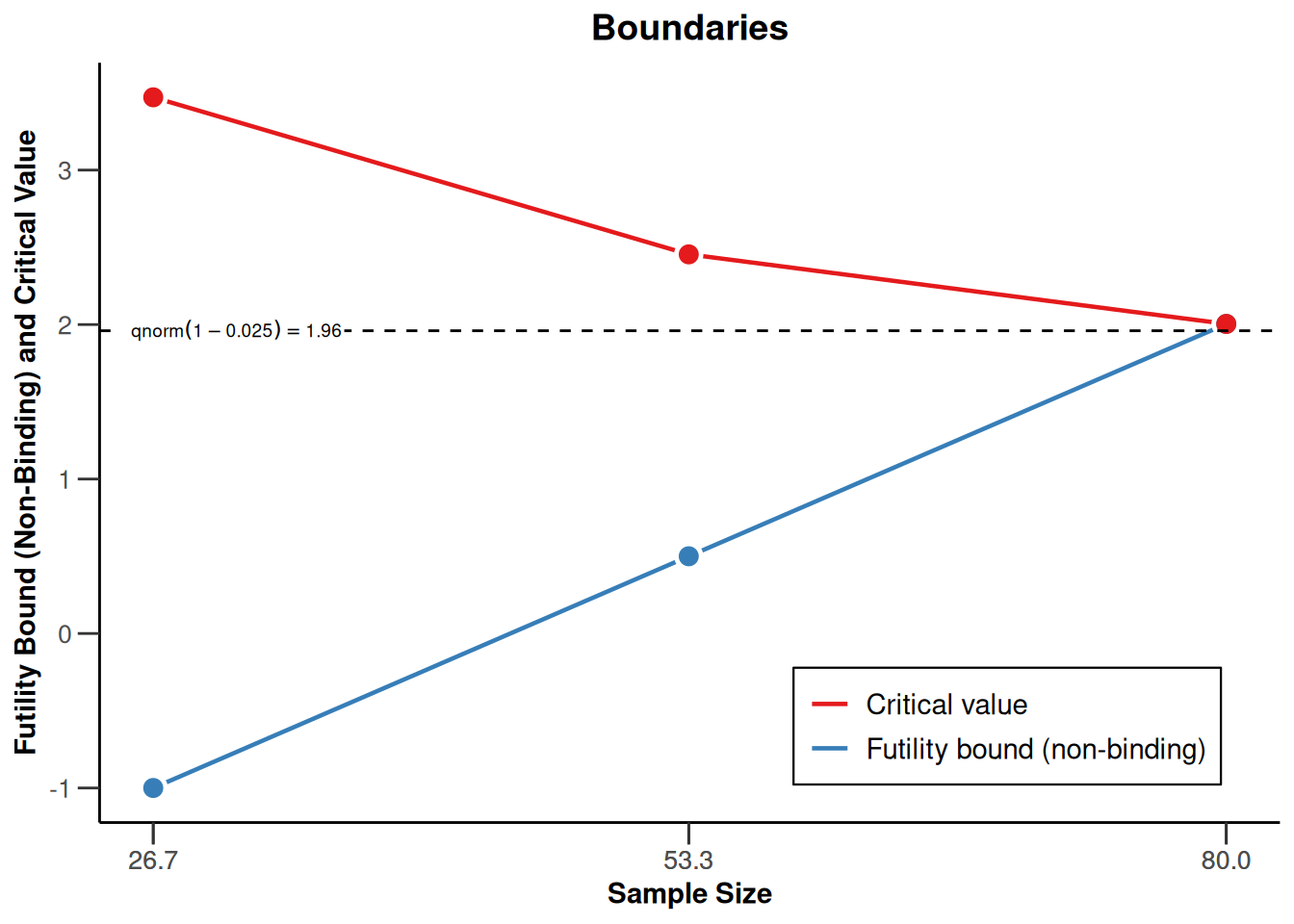
powerRates1 |> plot(type = 2)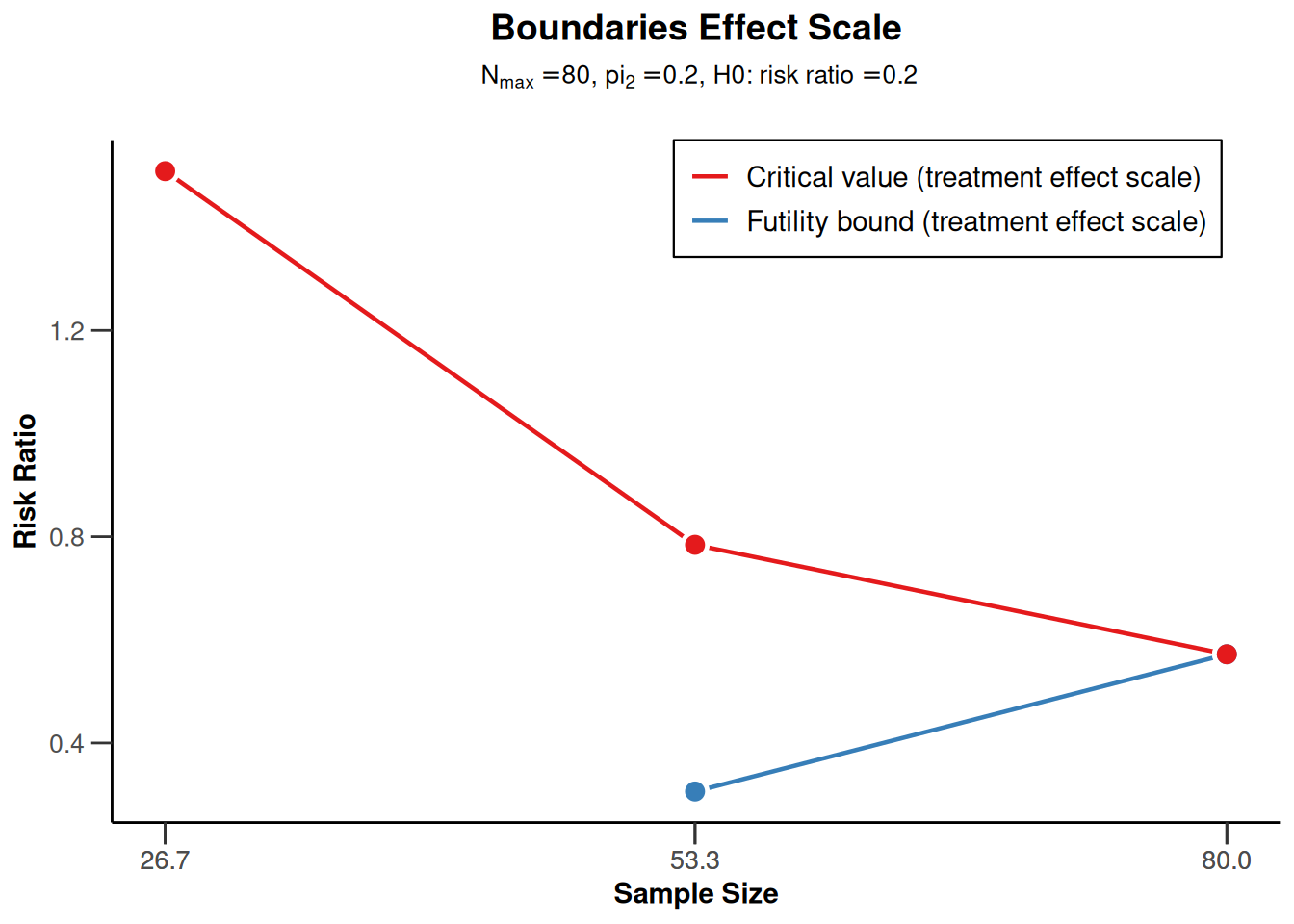
powerRates1 |> plot(type = 5)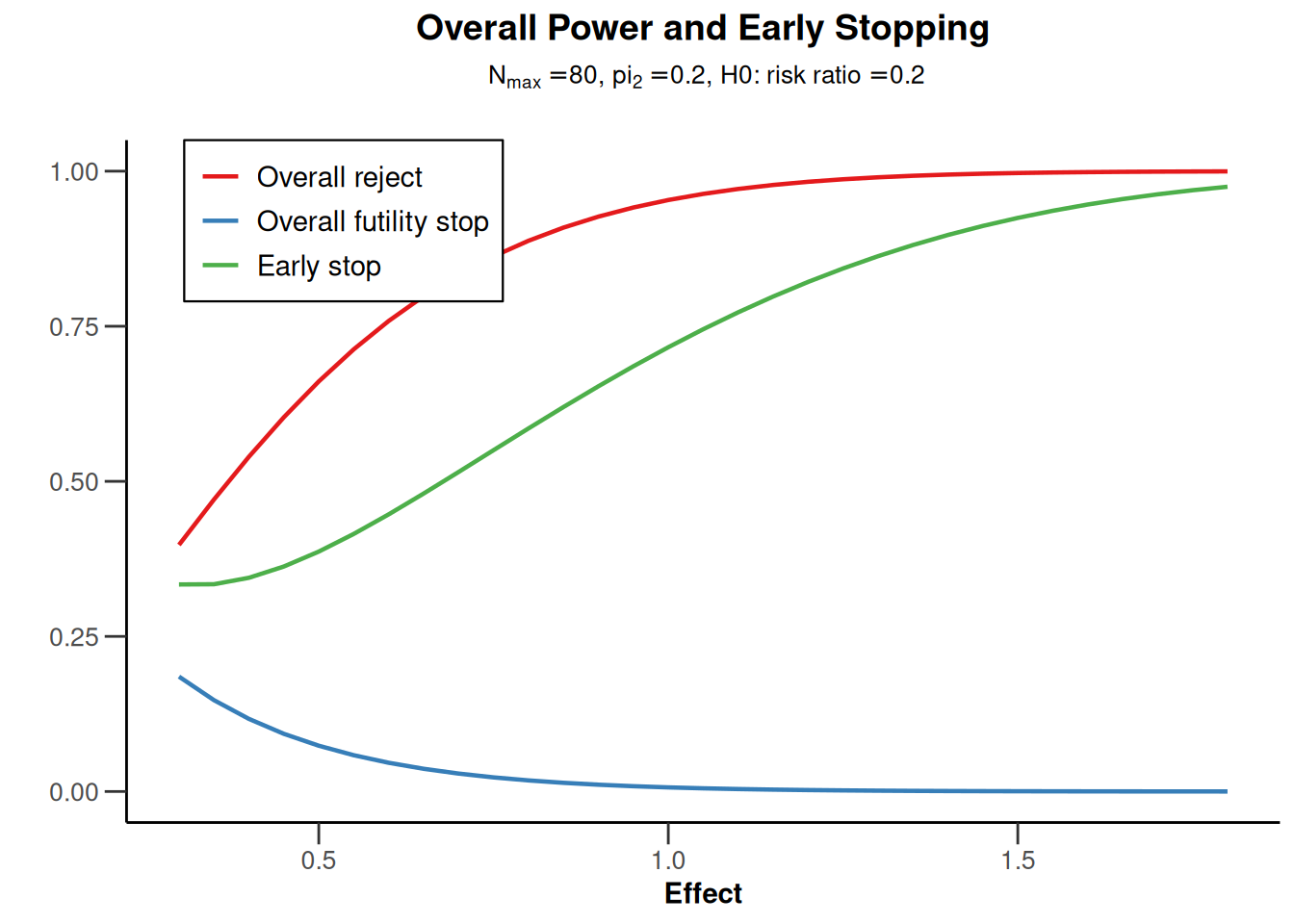
powerRates1 |> plot(type = 6)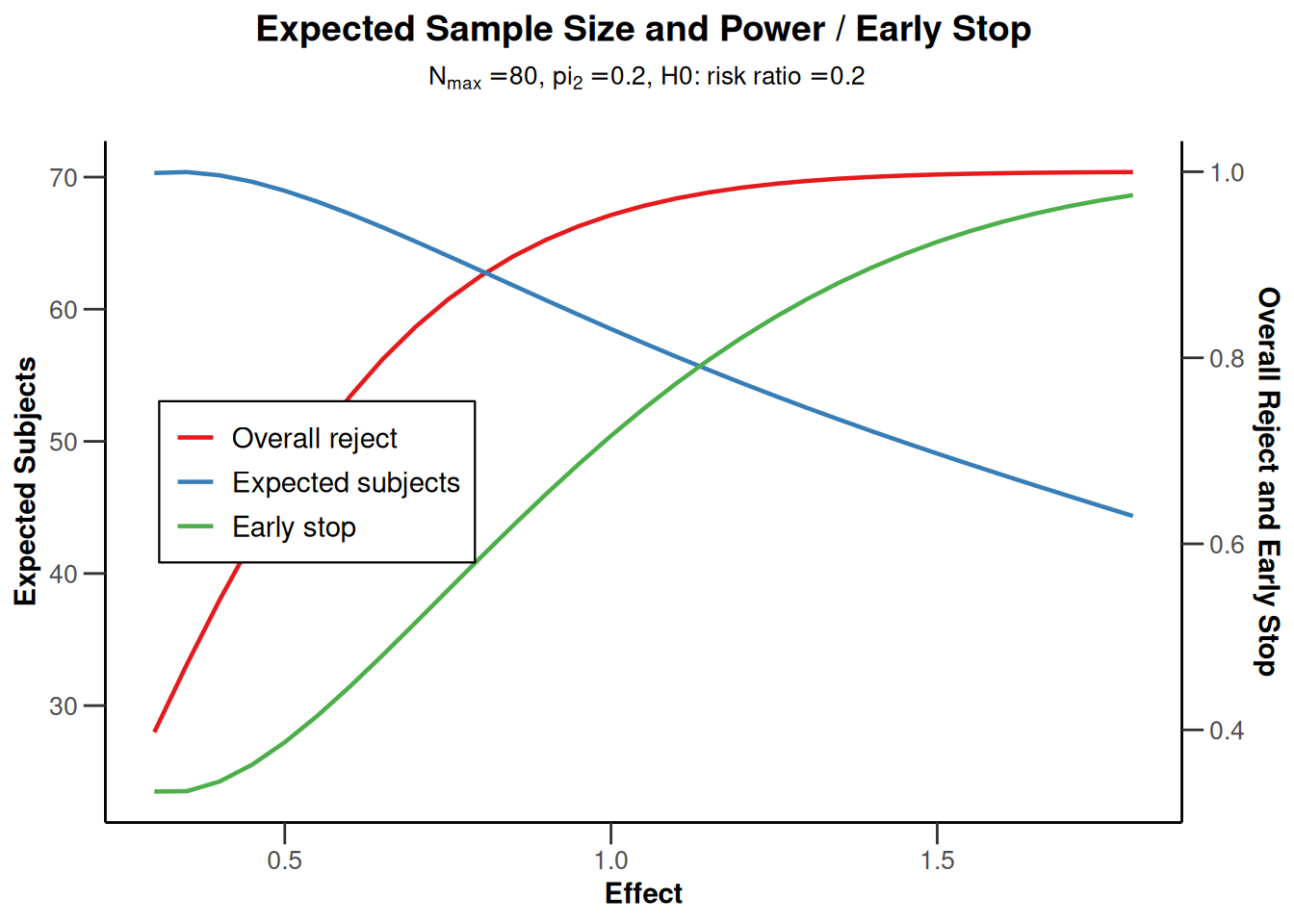
Power rates for a two-sided design
powerRates2 <- getDesignGroupSequential(
typeOfDesign = "OF",
sided = 2,
twoSidedPower = TRUE) |>
getPowerRates(maxNumberOfSubjects = 120)
powerRates2 |> plot(type = 1)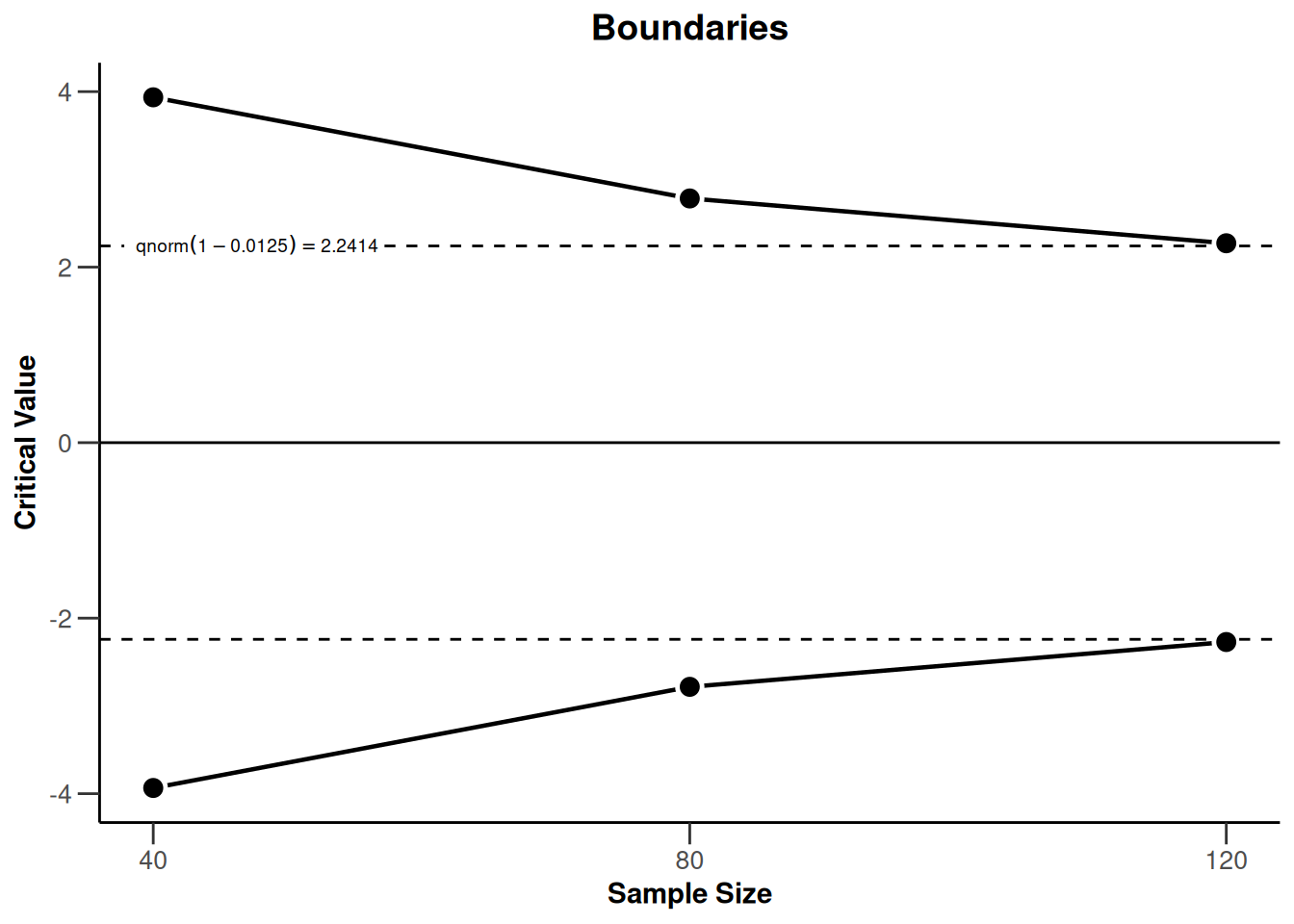
powerRates2 |> plot(type = 2)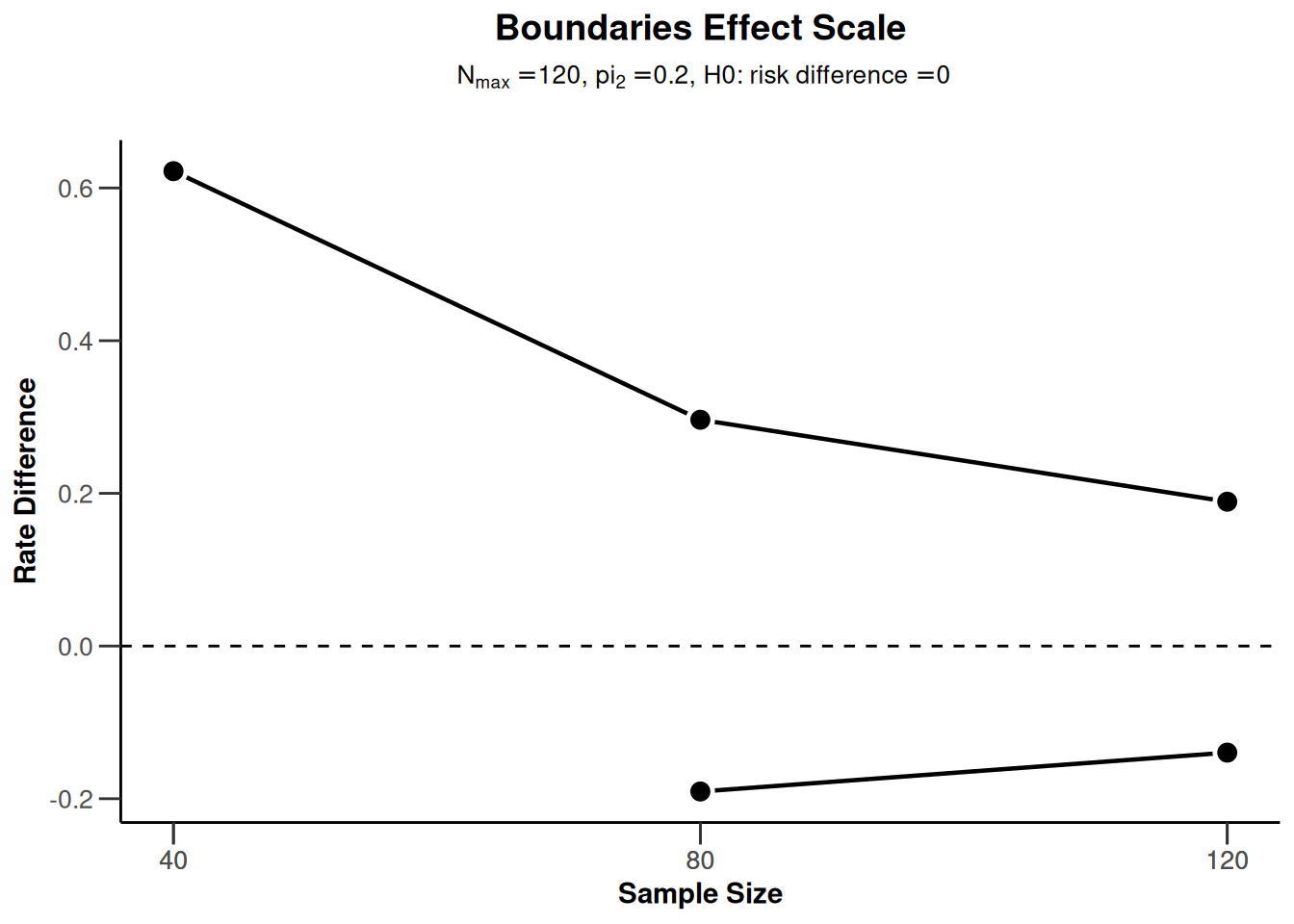
powerRates2 |> plot(type = 5)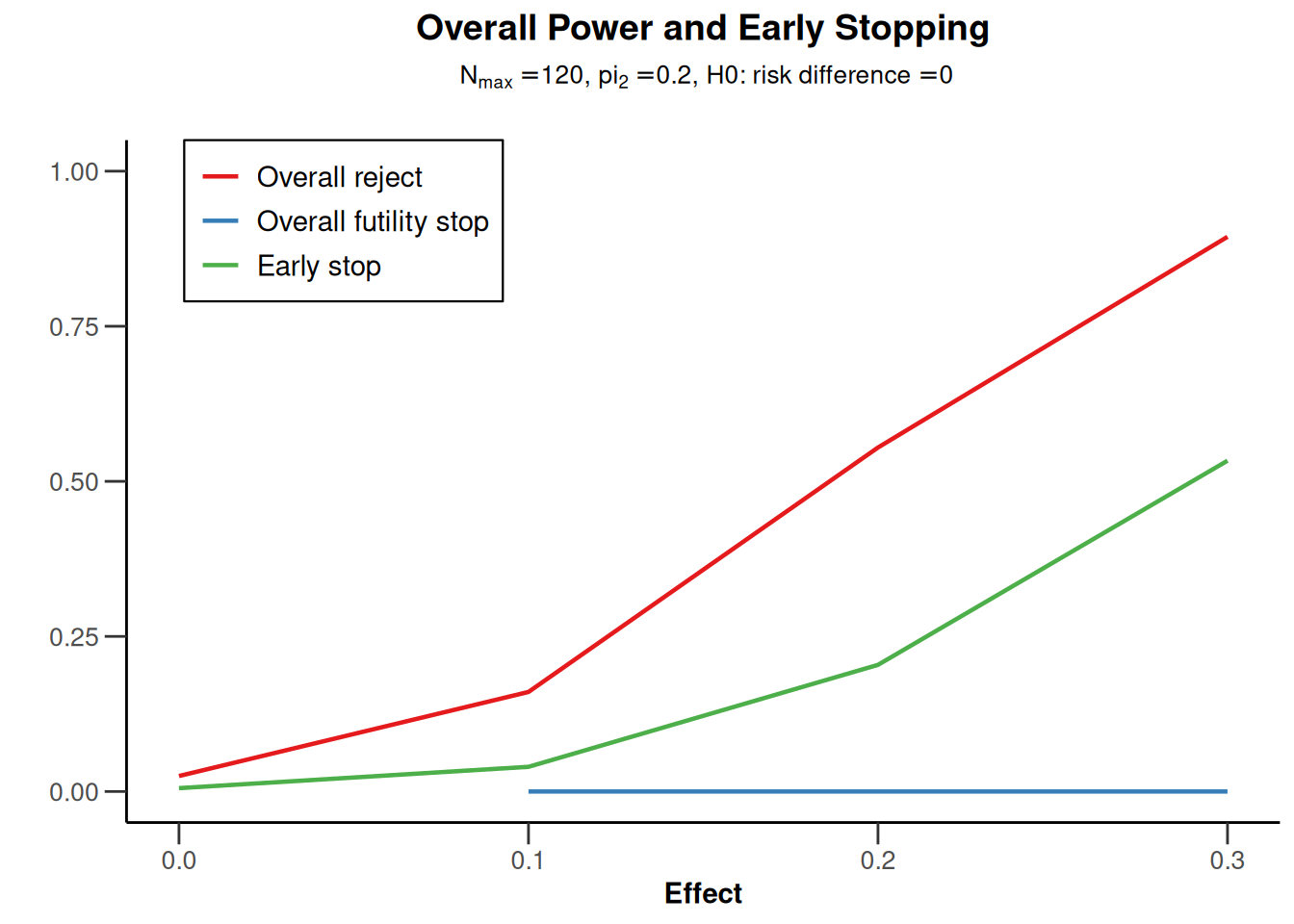
powerRates2 |> plot(type = 6)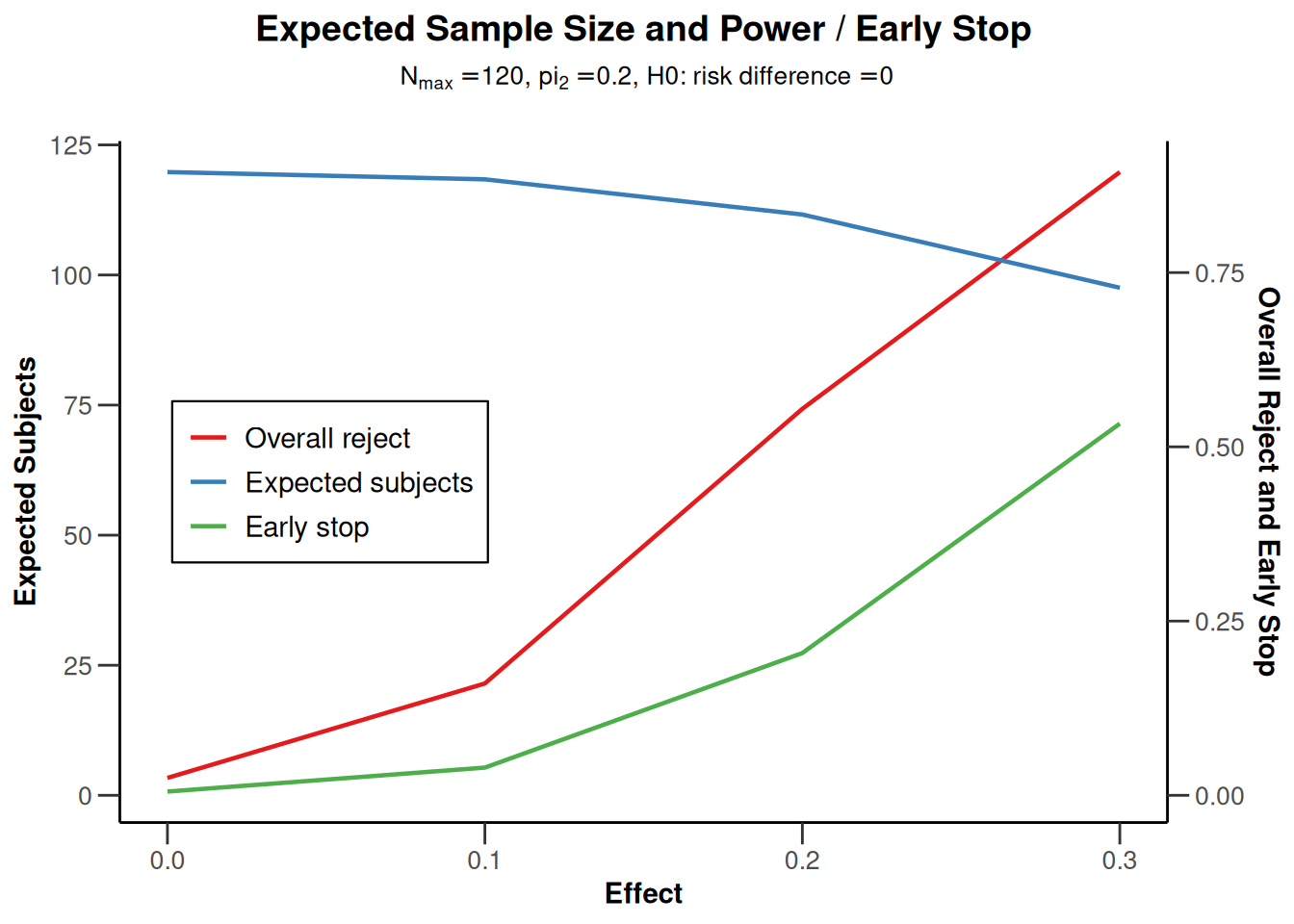
Power survival (survival endpoint)
Power survival for a one-sided design with futility bounds
powerSurvival <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(0, 0)) |>
getPowerSurvival(
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
maxNumberOfSubjects = 2480,
maxNumberOfEvents = 70
)
powerSurvival |> plot(type = 1)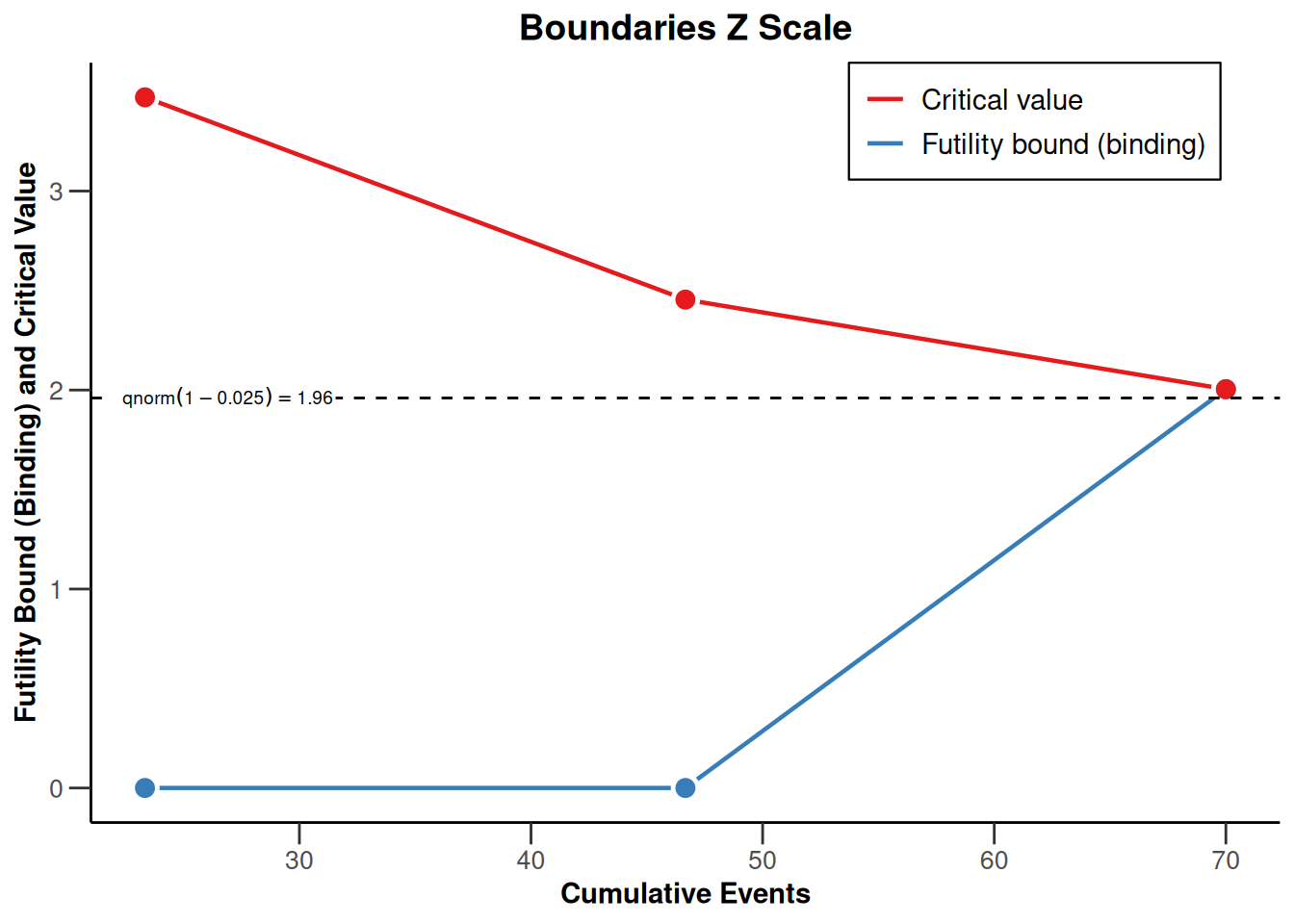
powerSurvival |> plot(type = 2)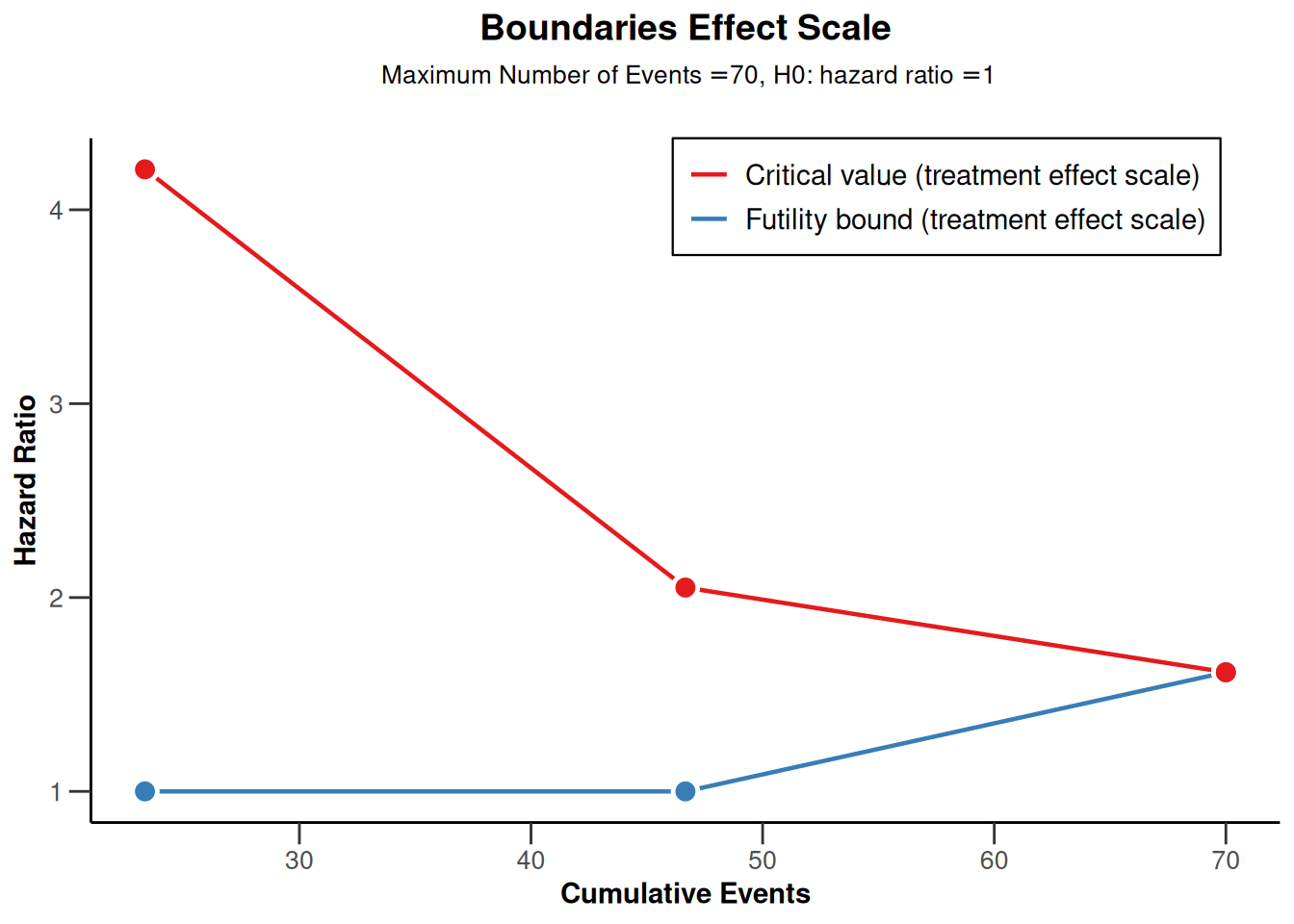
powerSurvival |> plot(type = 5)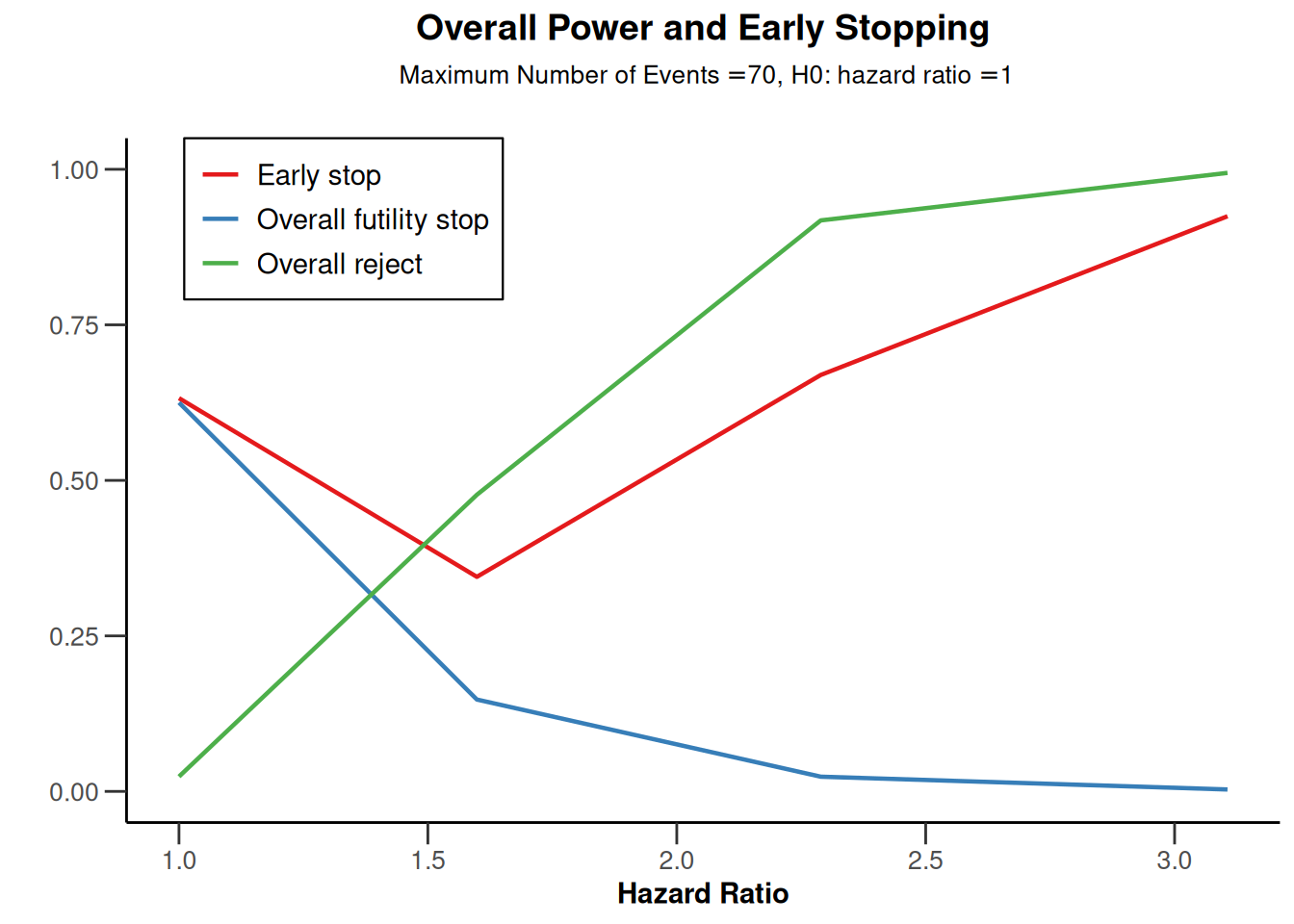
powerSurvival |> plot(type = 6)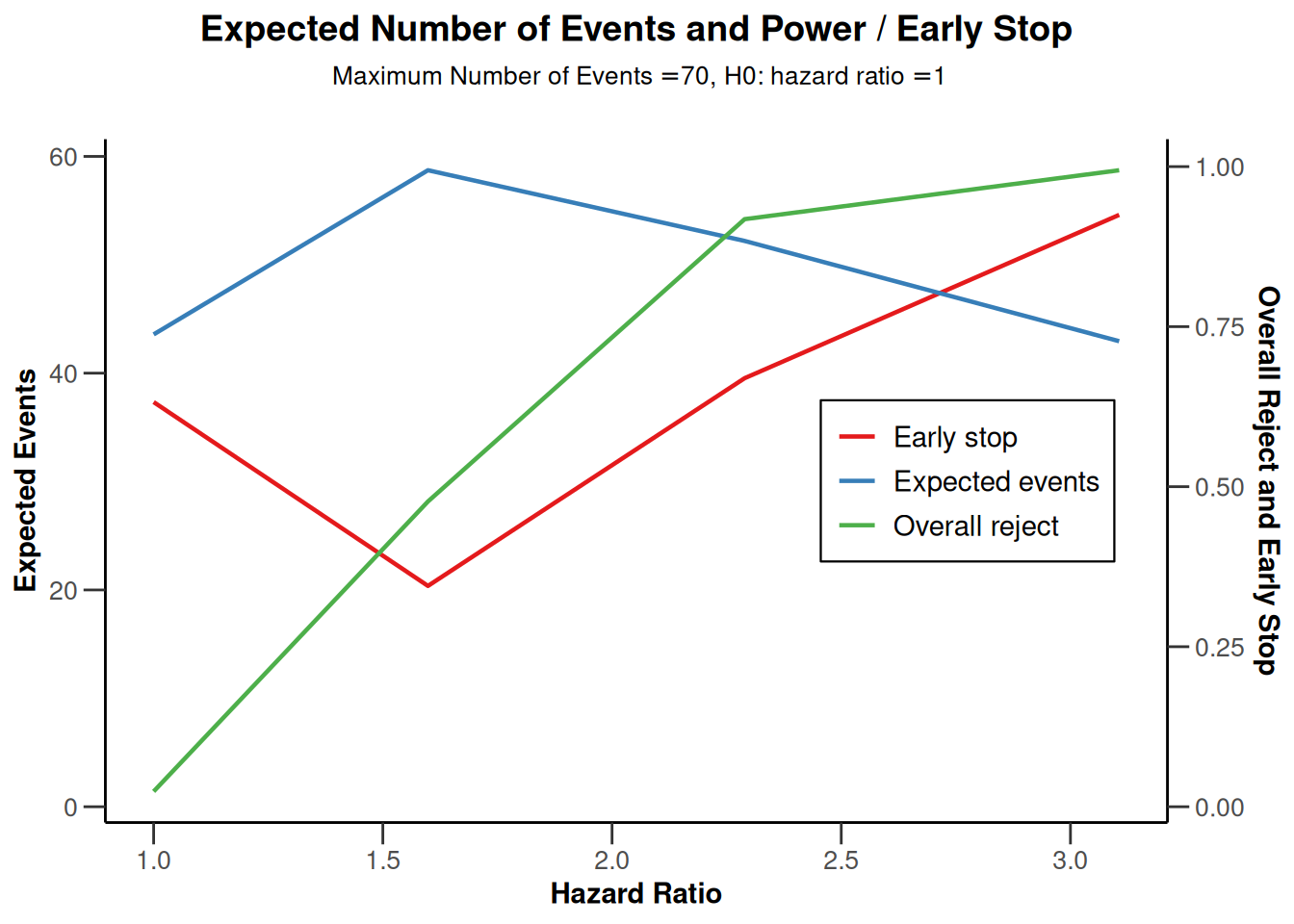
powerSurvival |> plot(type = 7)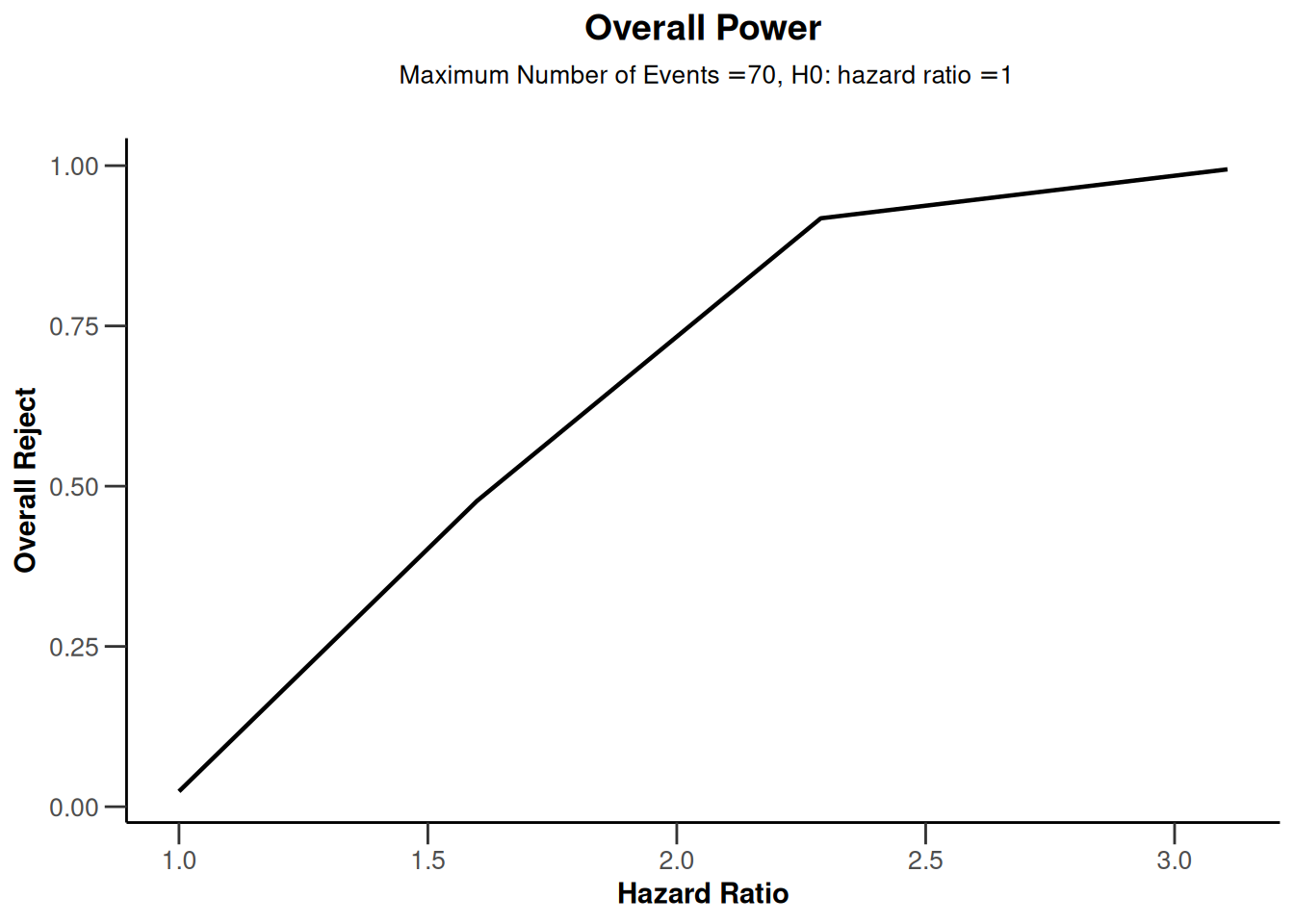
powerSurvival |> plot(type = 12)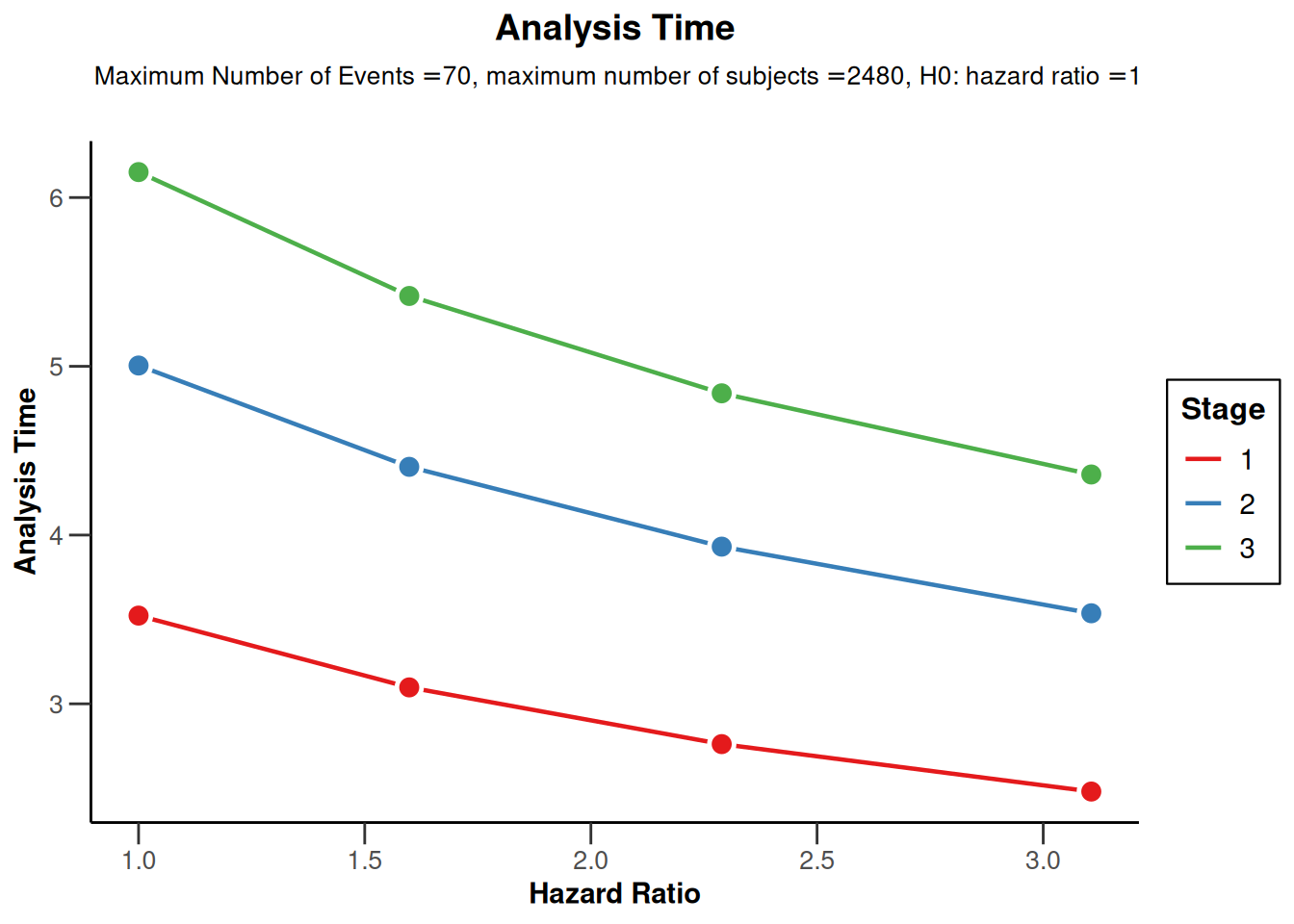
Power for a piecewise exponential survival distribution
piecewiseSurvivalTime <- list(
"<5" = 0.04,
"5 - <10" = 0.02,
">= 10" = 0.008
)
powerSurvival1 <- getDesignGroupSequential(
kMax = 3,
typeOfDesign = "OF",
sided = 2,
twoSidedPower = TRUE
) |>
getPowerSurvival(
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
piecewiseSurvivalTime = piecewiseSurvivalTime,
maxNumberOfSubjects = 2480,
maxNumberOfEvents = 70,
hazardRatio = c(0.5, 2)
)
powerSurvival1 |> plot(type = 1)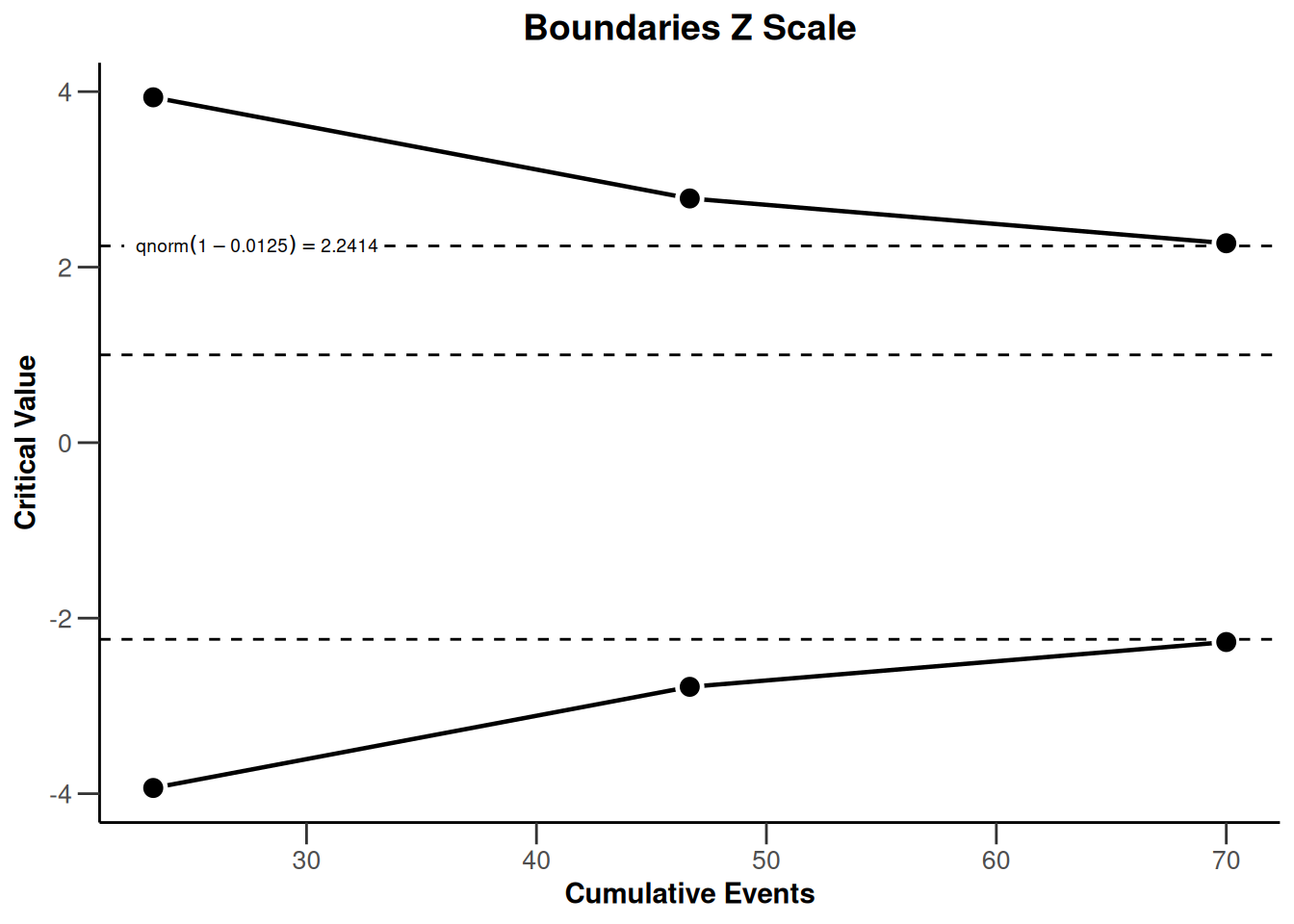
powerSurvival1 |> plot(type = 2)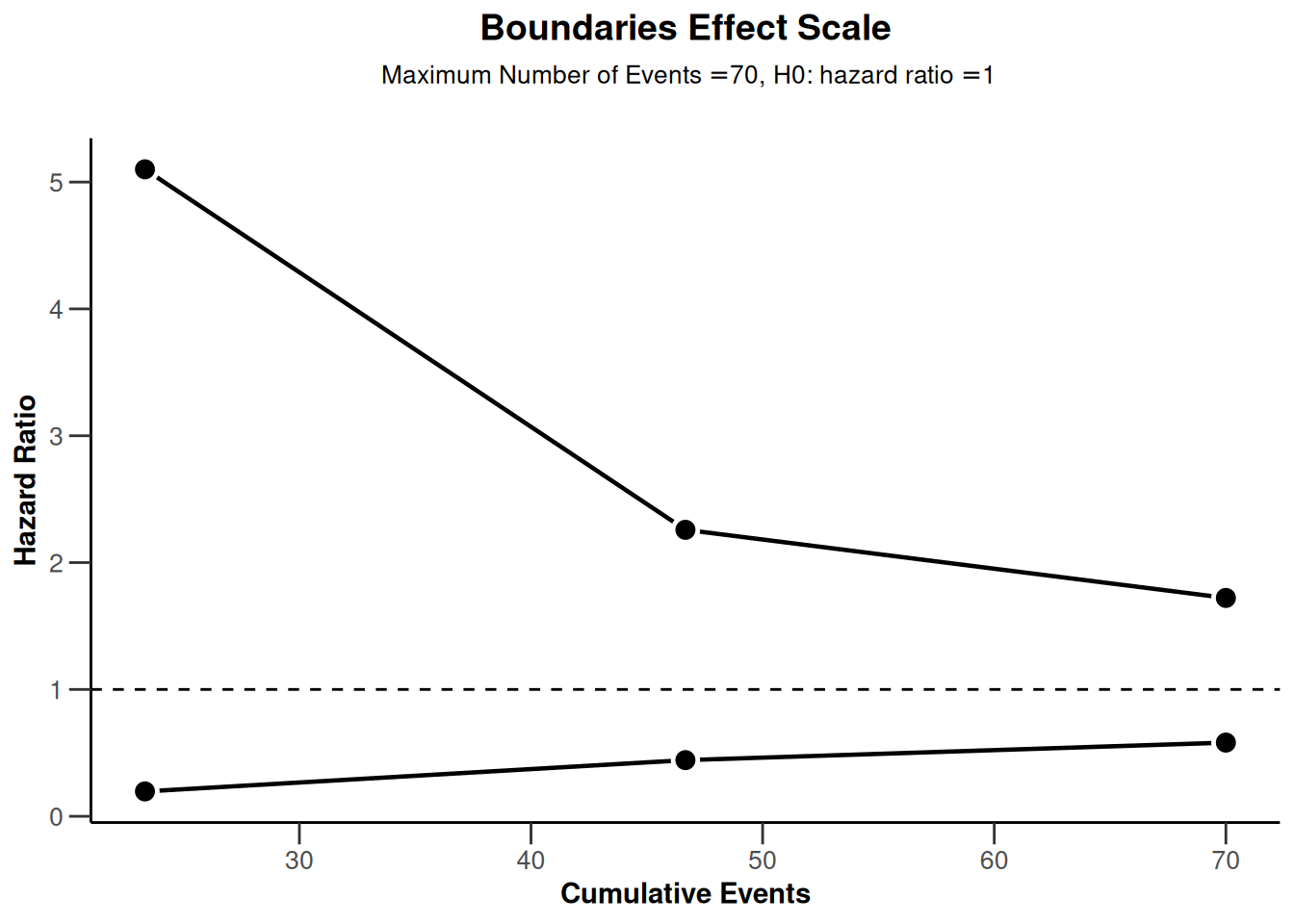
powerSurvival1 |> plot(type = 12)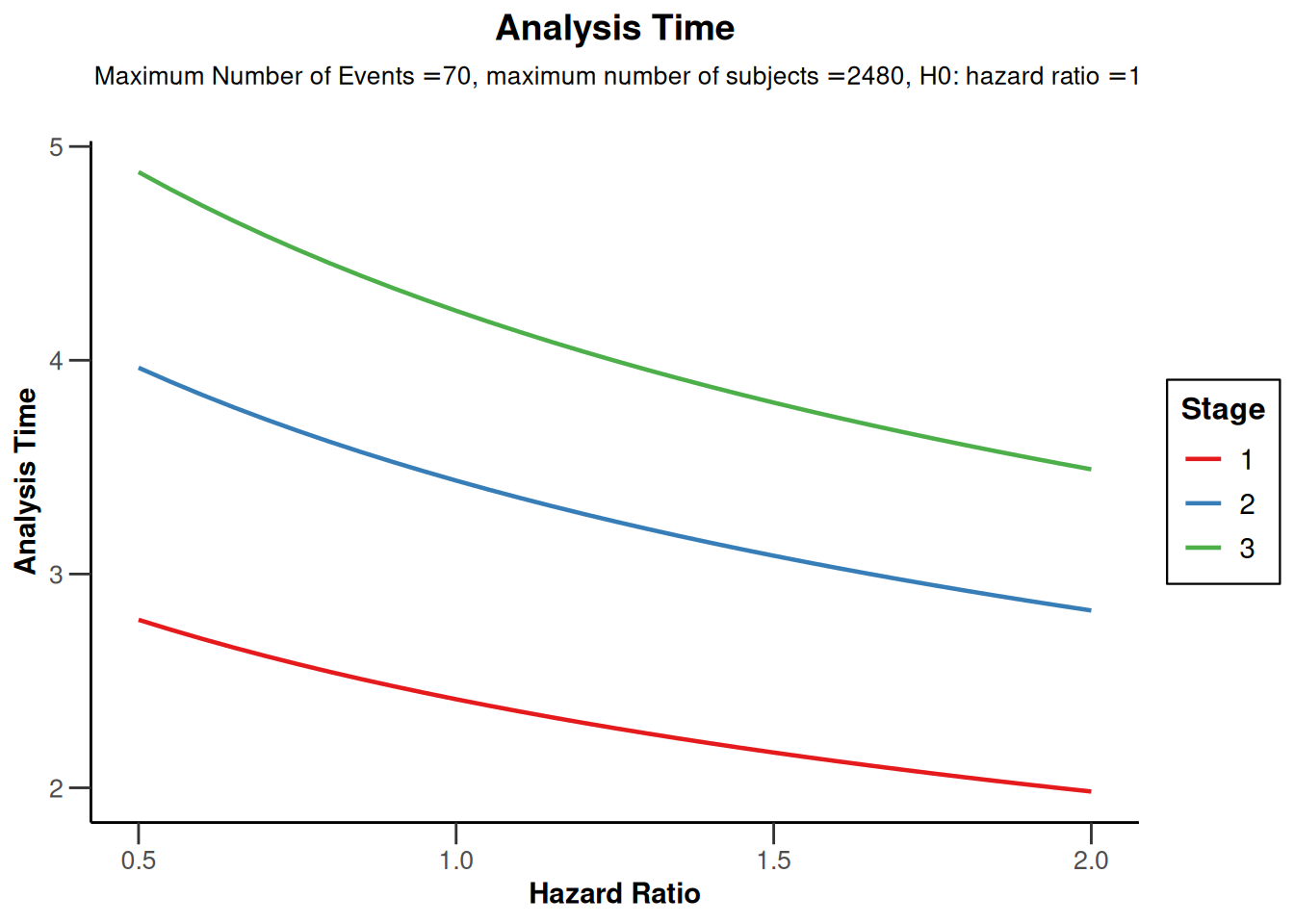
powerSurvival1 |> plot(type = 13, legendPosition = 1)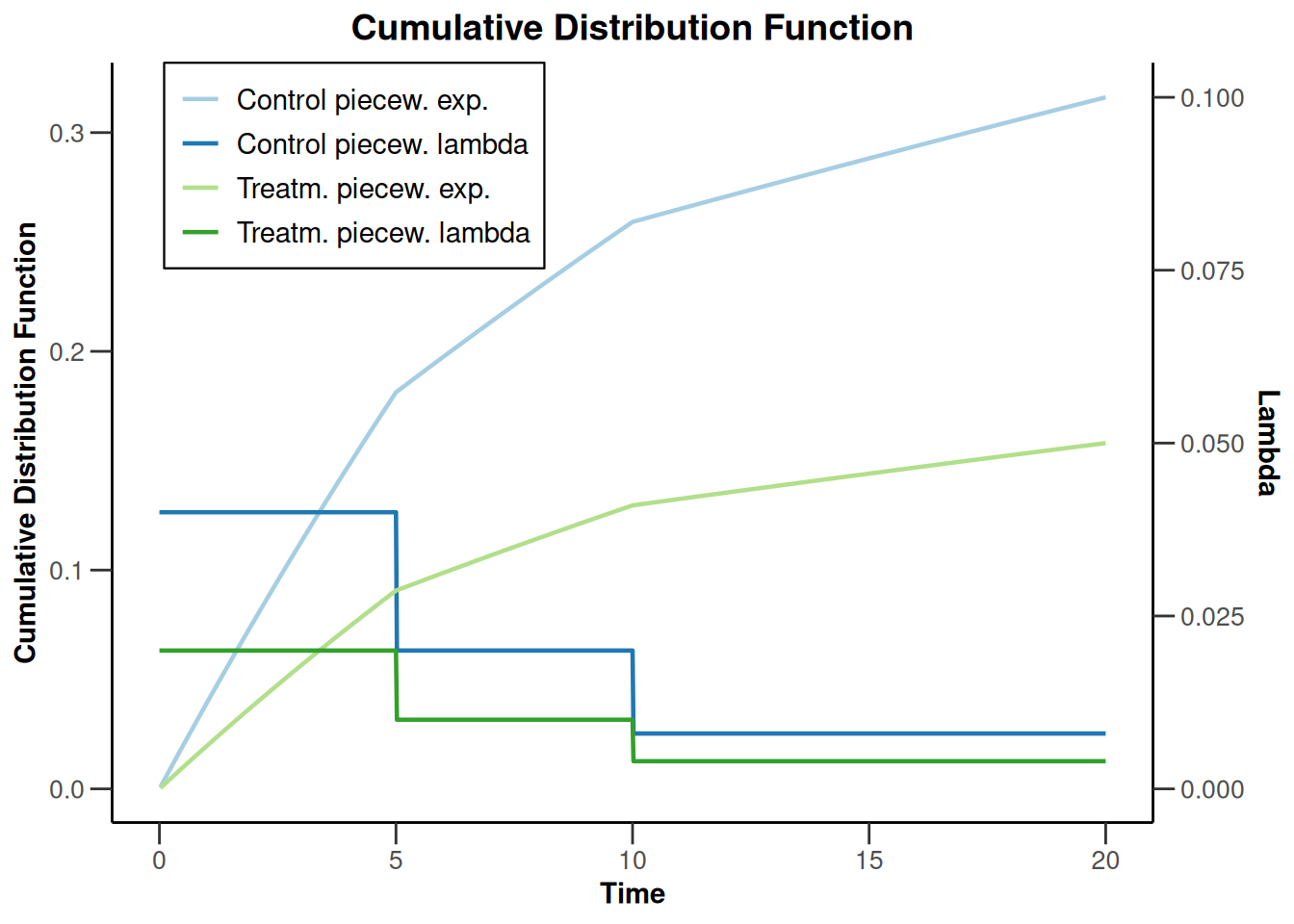
powerSurvival1 |> plot(type = 14, legendPosition = 5)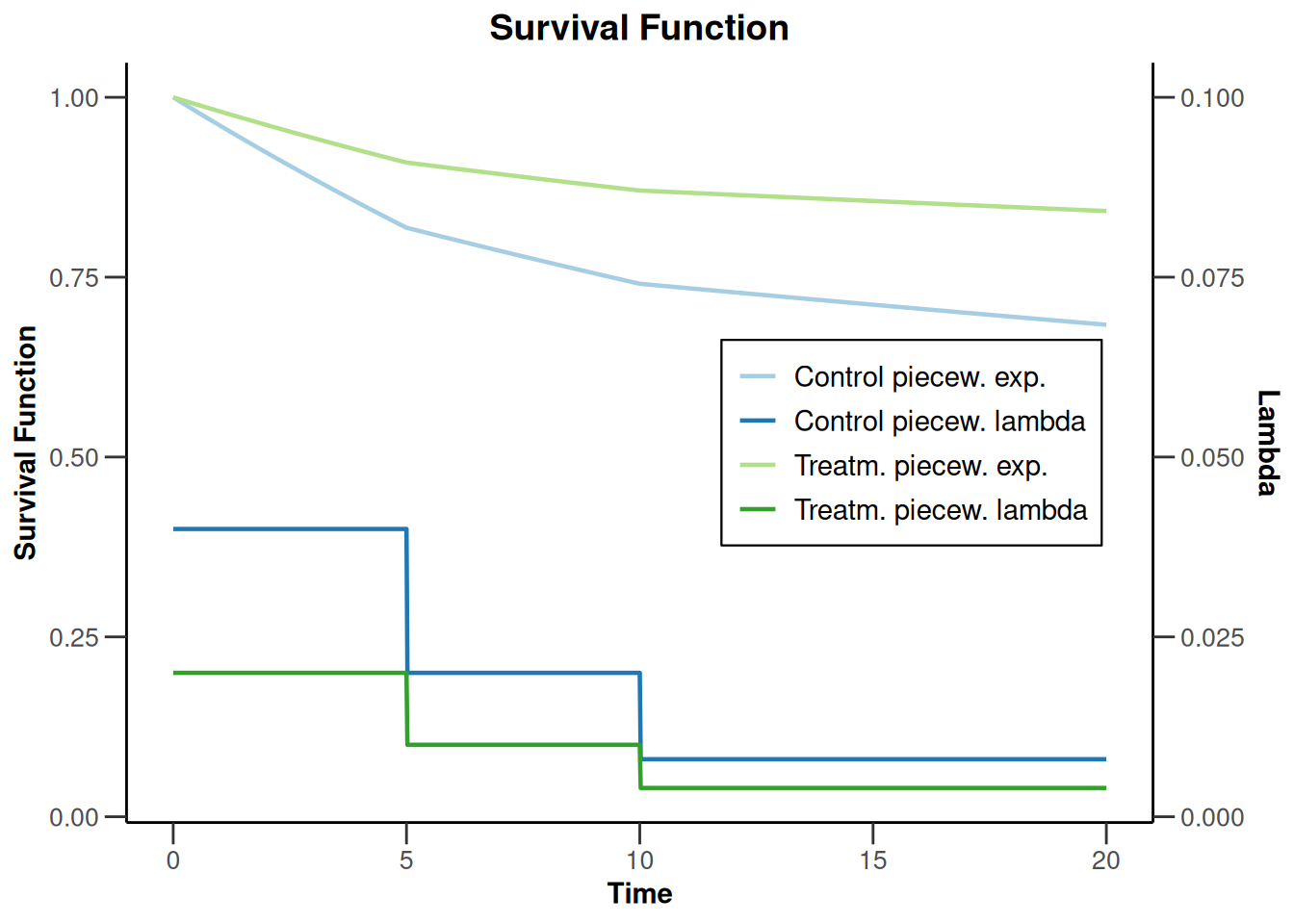
Power for a piecewise exponential survival distribution and a design with futility bounds
piecewiseSurvivalTime <- list(
"0 - <6" = 0.025,
"6 - <9" = 0.04,
"9 - <15" = 0.015,
"15 - <21" = 0.01,
">= 21" = 0.007
)
powerSurvival2 <- getDesignGroupSequential(
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(0, 0.1)) |>
getPowerSurvival(
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
piecewiseSurvivalTime = piecewiseSurvivalTime,
maxNumberOfSubjects = 2480,
maxNumberOfEvents = 70,
hazardRatio = 0.8
)
powerSurvival2 |> plot(type = 1)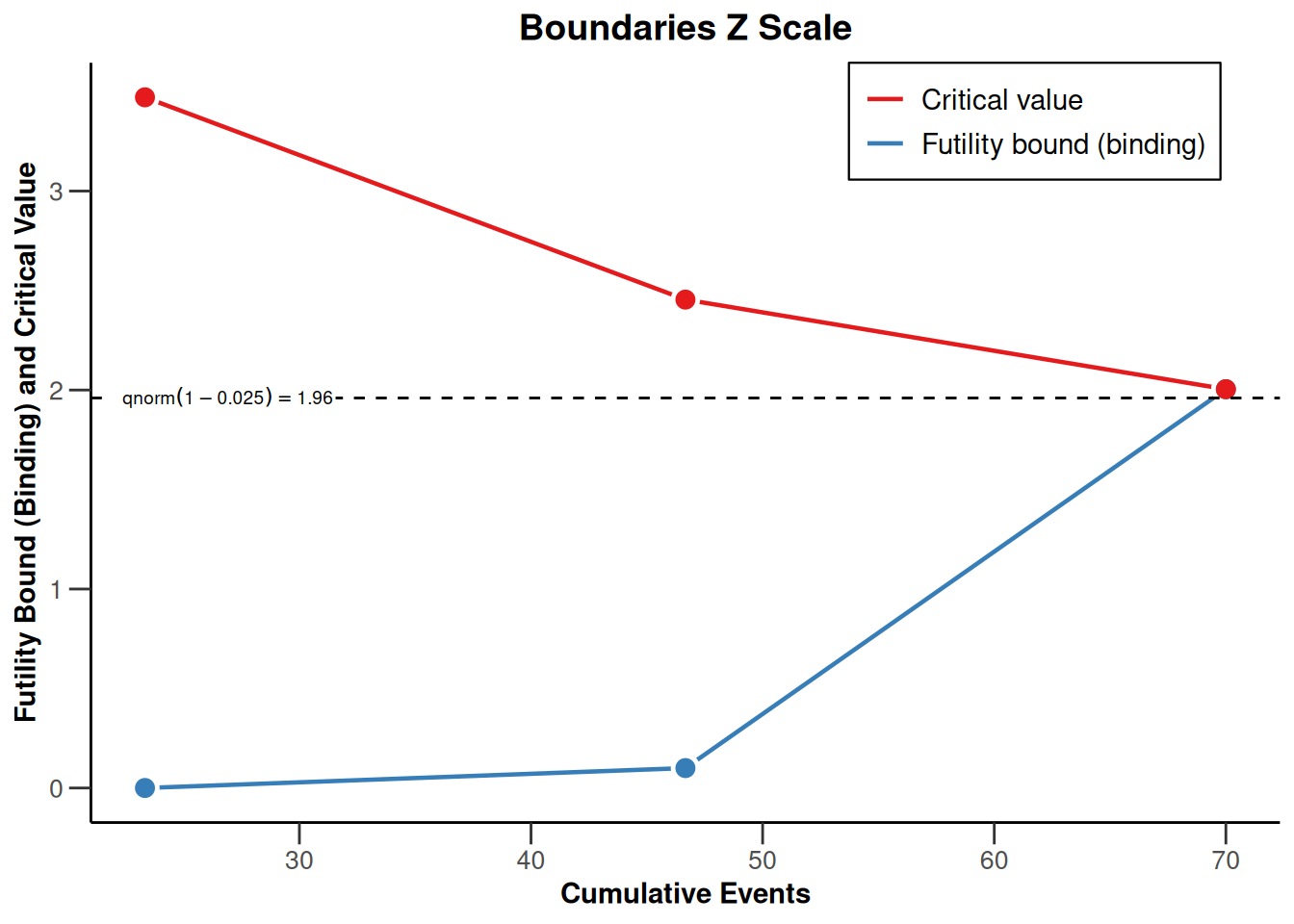
powerSurvival2 |> plot(type = 2)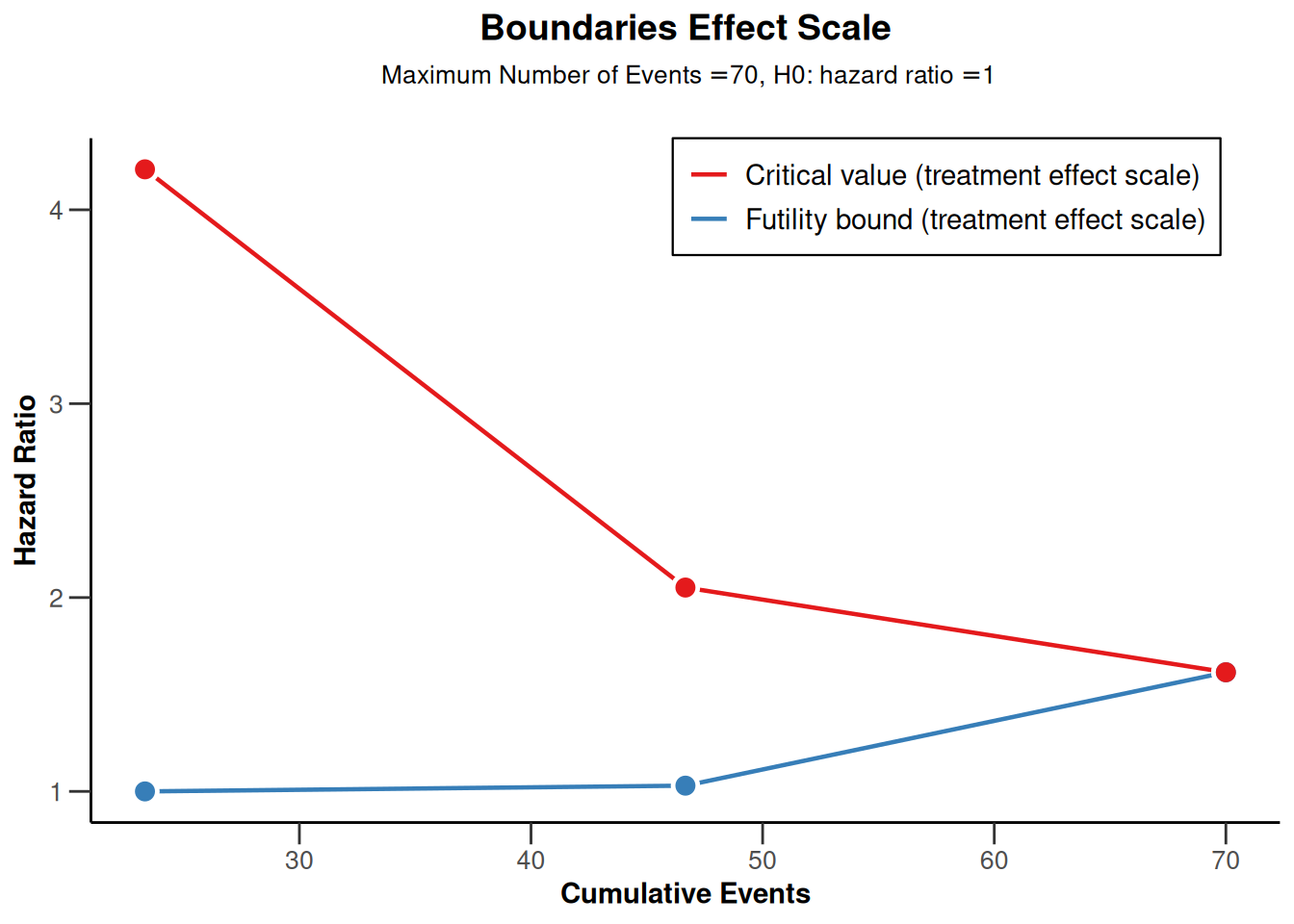
powerSurvival2 |> plot(type = 13, legendPosition = 1)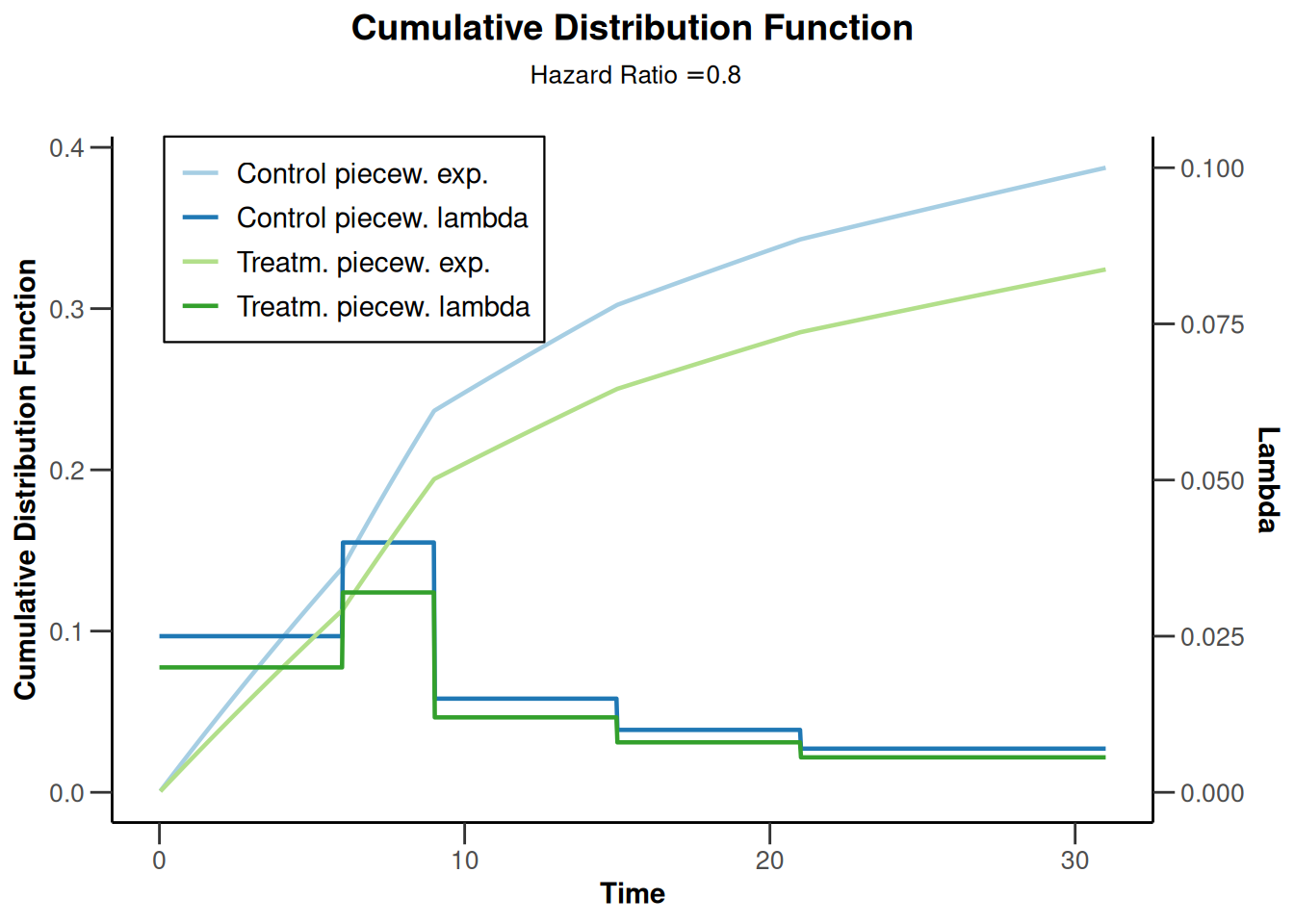
powerSurvival2 |> plot(type = 14)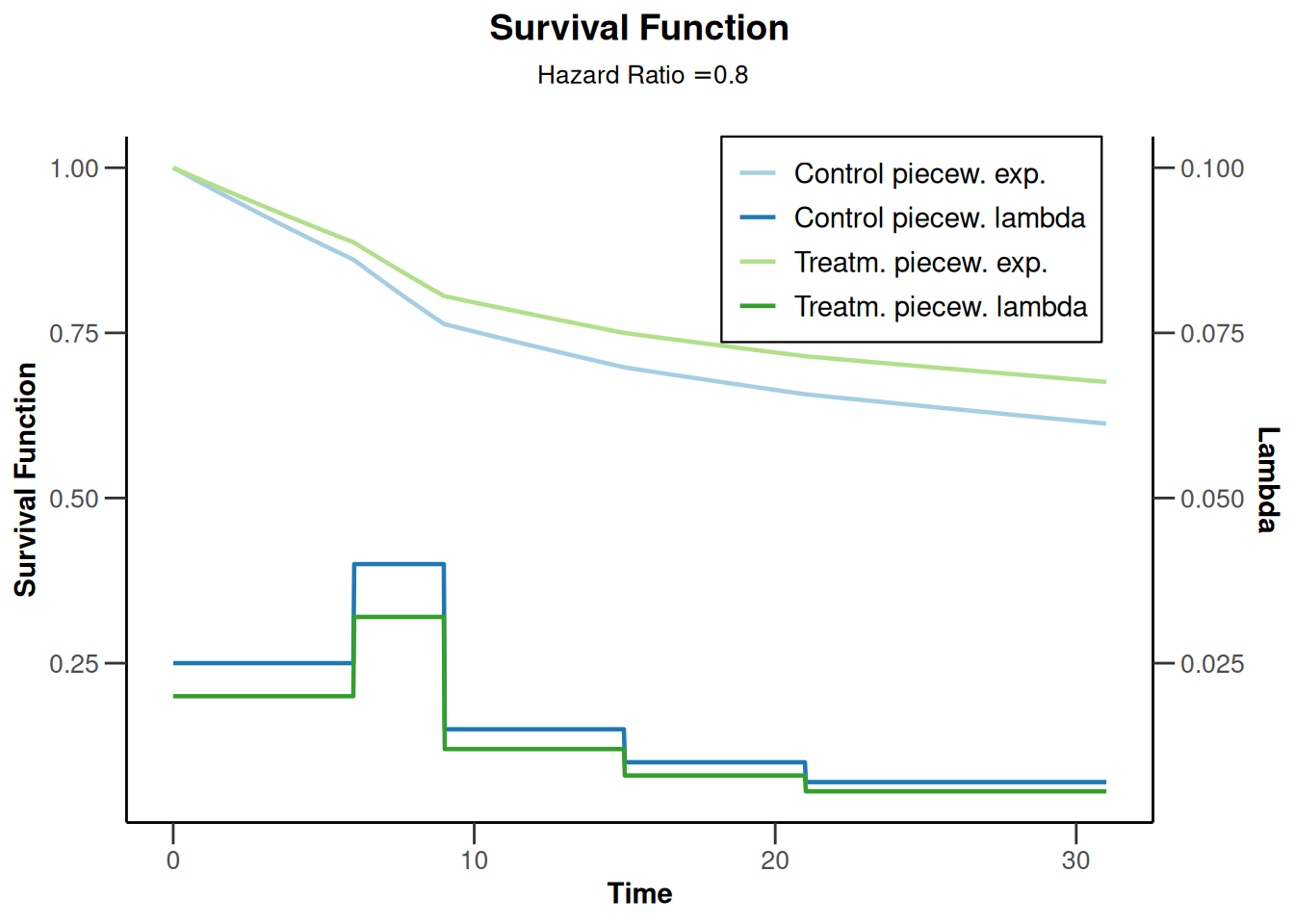
powerSurvival3 <- getDesignGroupSequential(
typeOfDesign = "OF",
sided = 1,
futilityBounds = c(0, 0.1)) |>
getPowerSurvival(
typeOfComputation = "Schoenfeld",
thetaH0 = 1,
allocationRatioPlanned = 1,
kappa = 1,
piecewiseSurvivalTime = c(0, 5, 10),
lambda2 = c(0.025, 0.04, 0.015),
lambda1 = c(0.02, 0.032, 0.012),
maxNumberOfSubjects = 2480,
maxNumberOfEvents = 70
)
powerSurvival3 |> plot(type = 1)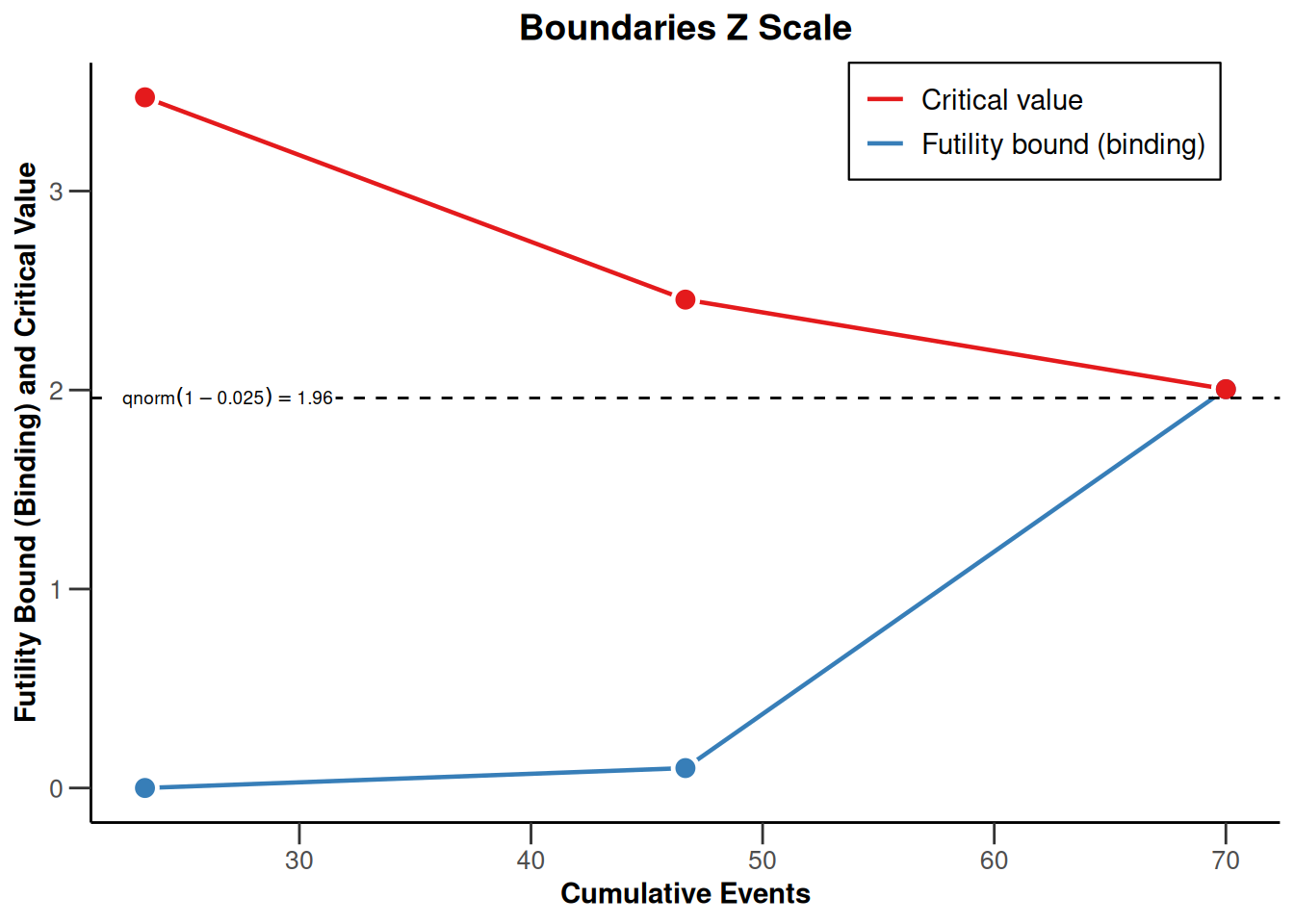
powerSurvival3 |> plot(type = 2)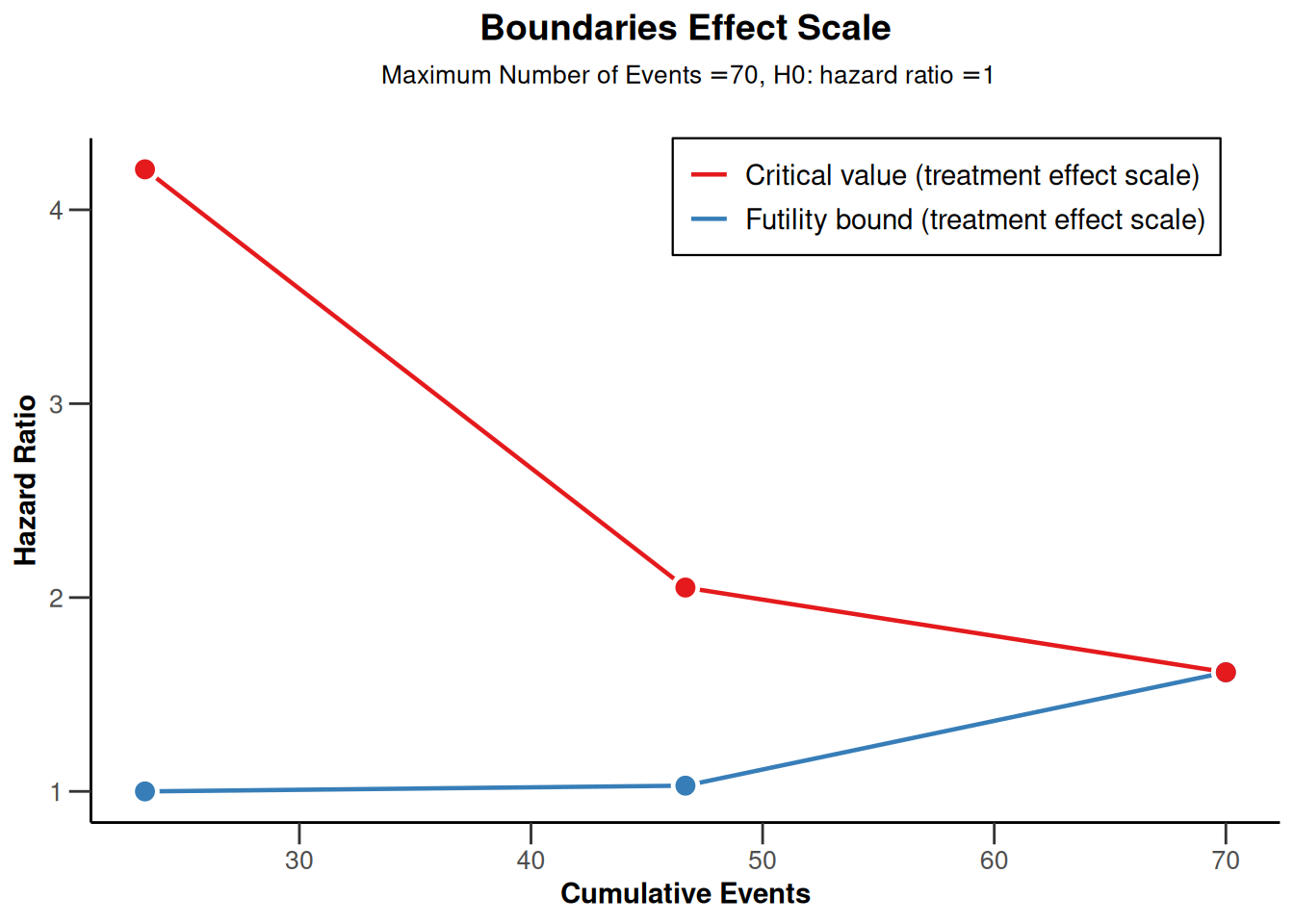
powerSurvival3 |> plot(type = 13, legendPosition = 1)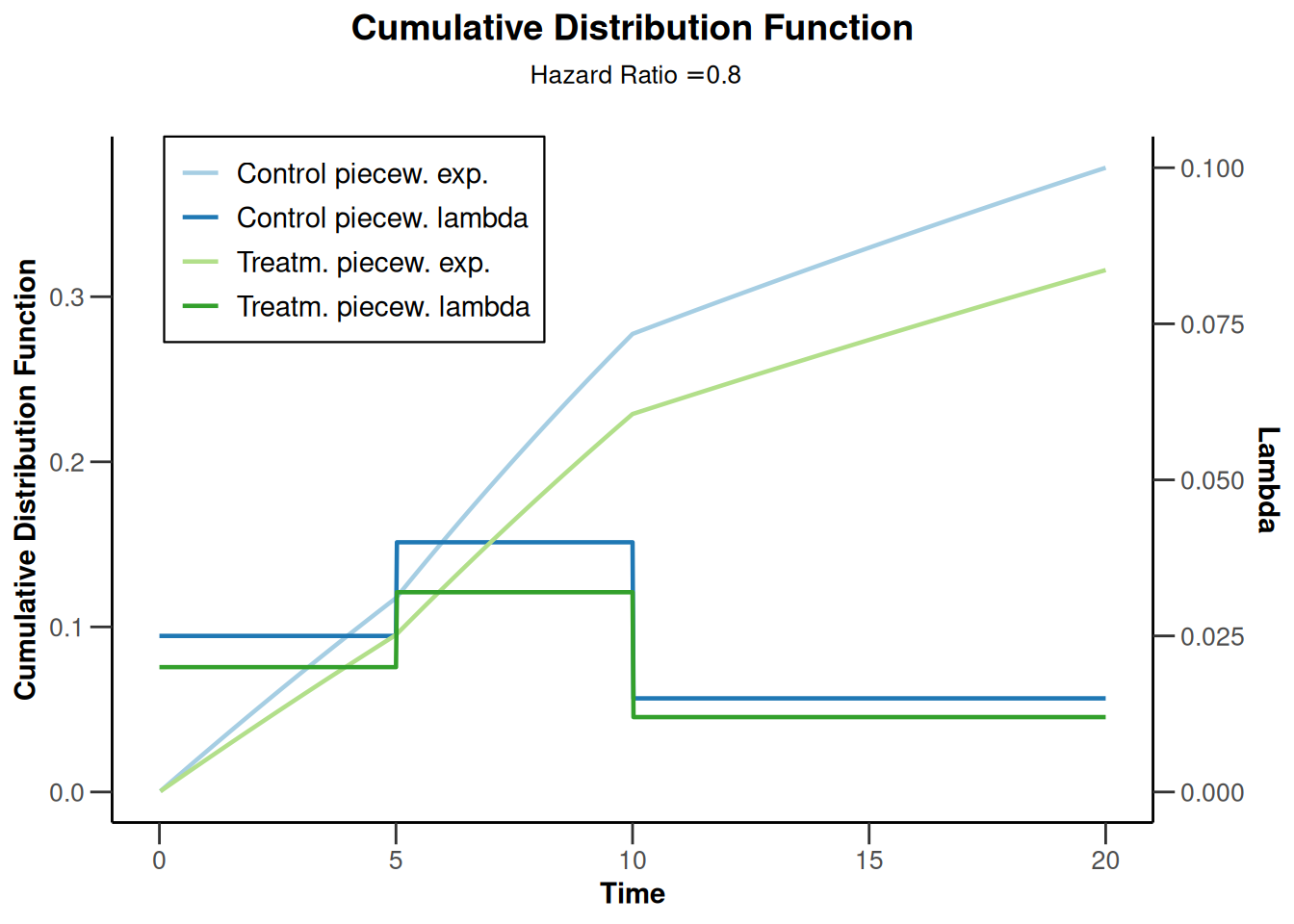
powerSurvival3 |> plot(type = 14)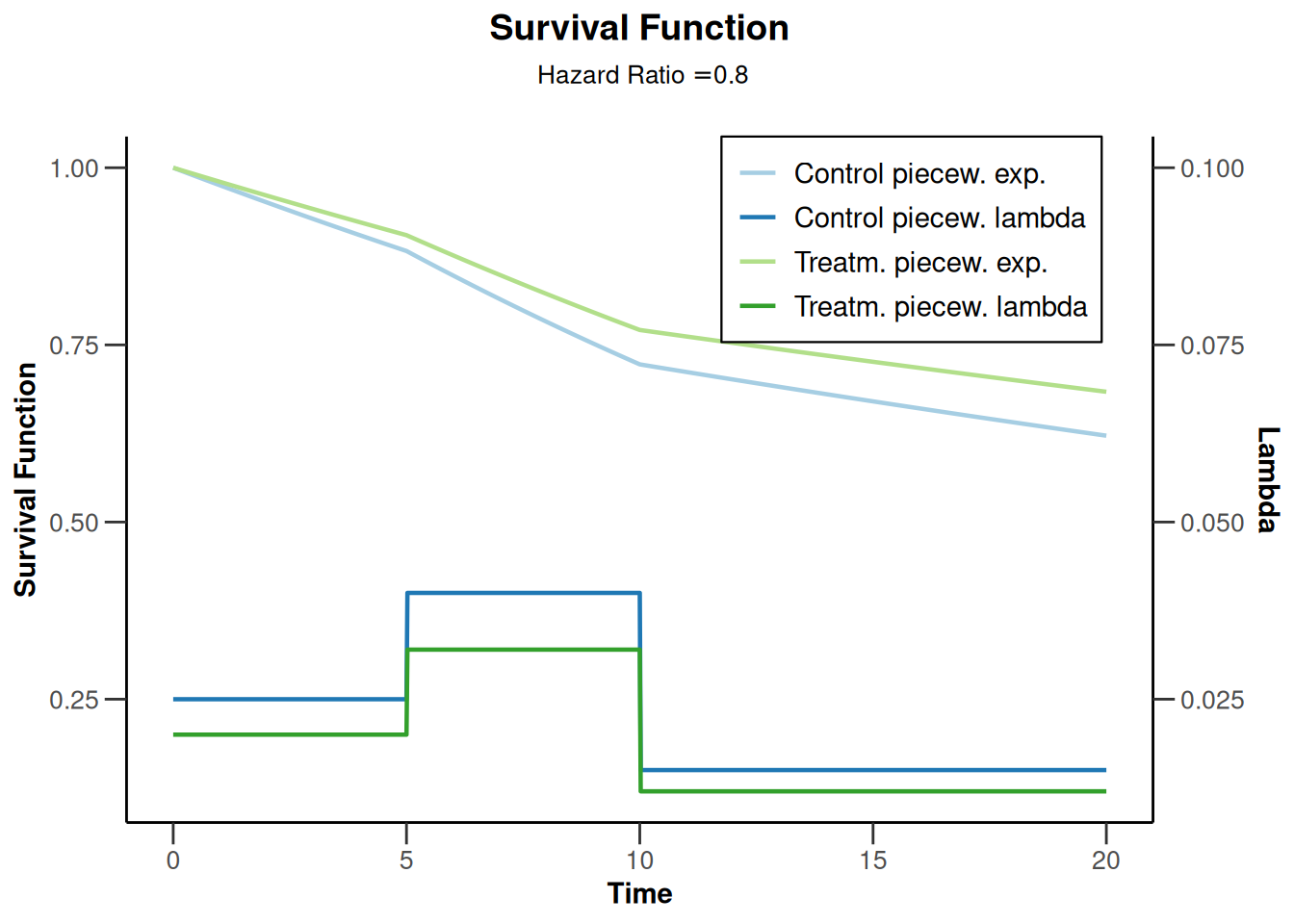
Power survival for one lambda
powerSurvival4 <- getPowerSurvival(
accrualTime = 12,
lambda2 = 0.04,
hazardRatio = 0.6,
maxNumberOfSubjects = 1400,
maxNumberOfEvents = 300
)
powerSurvival4 |> plot(type = 13, legendPosition = 1)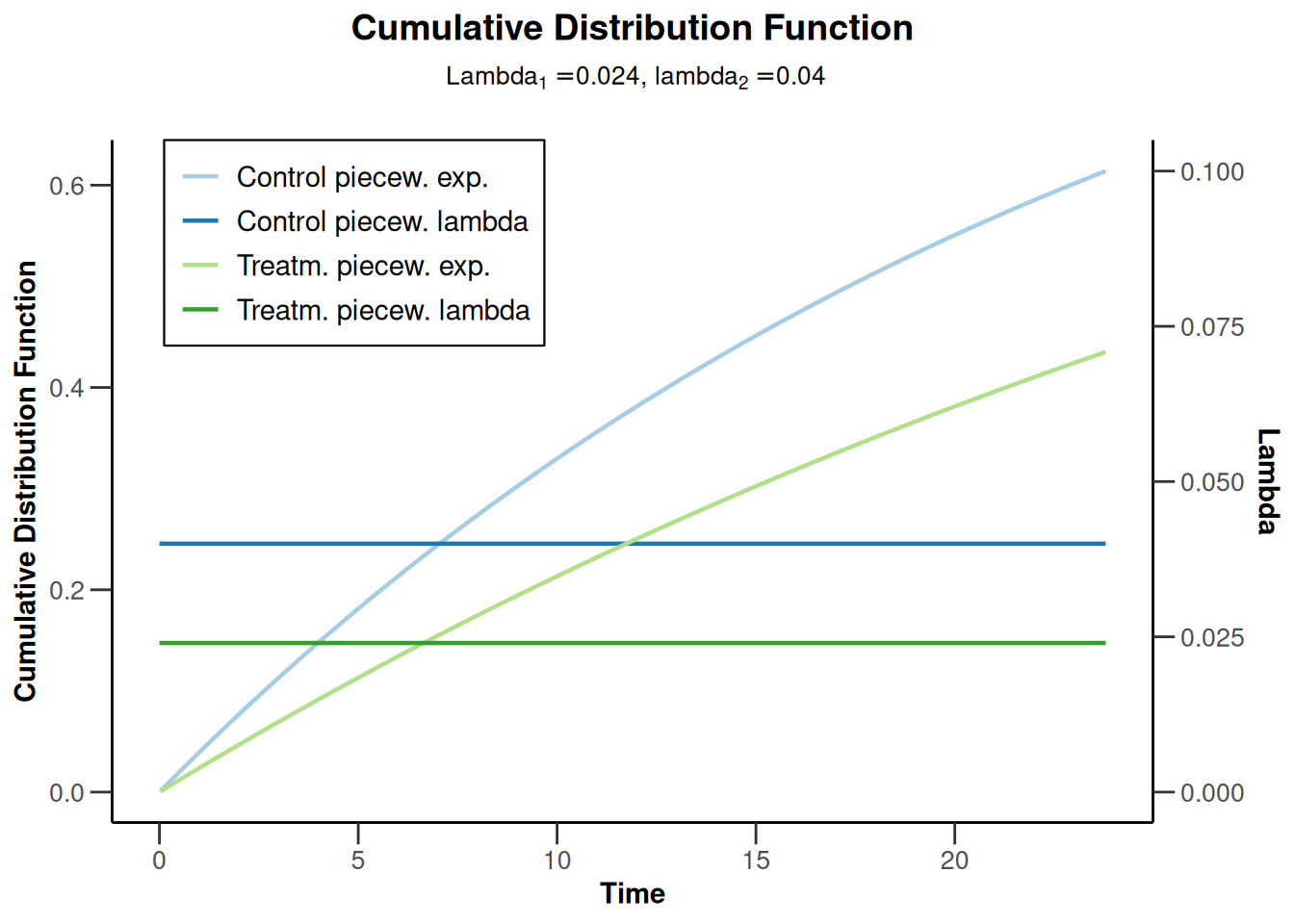
powerSurvival4 |> plot(type = 14, legendPosition = 5)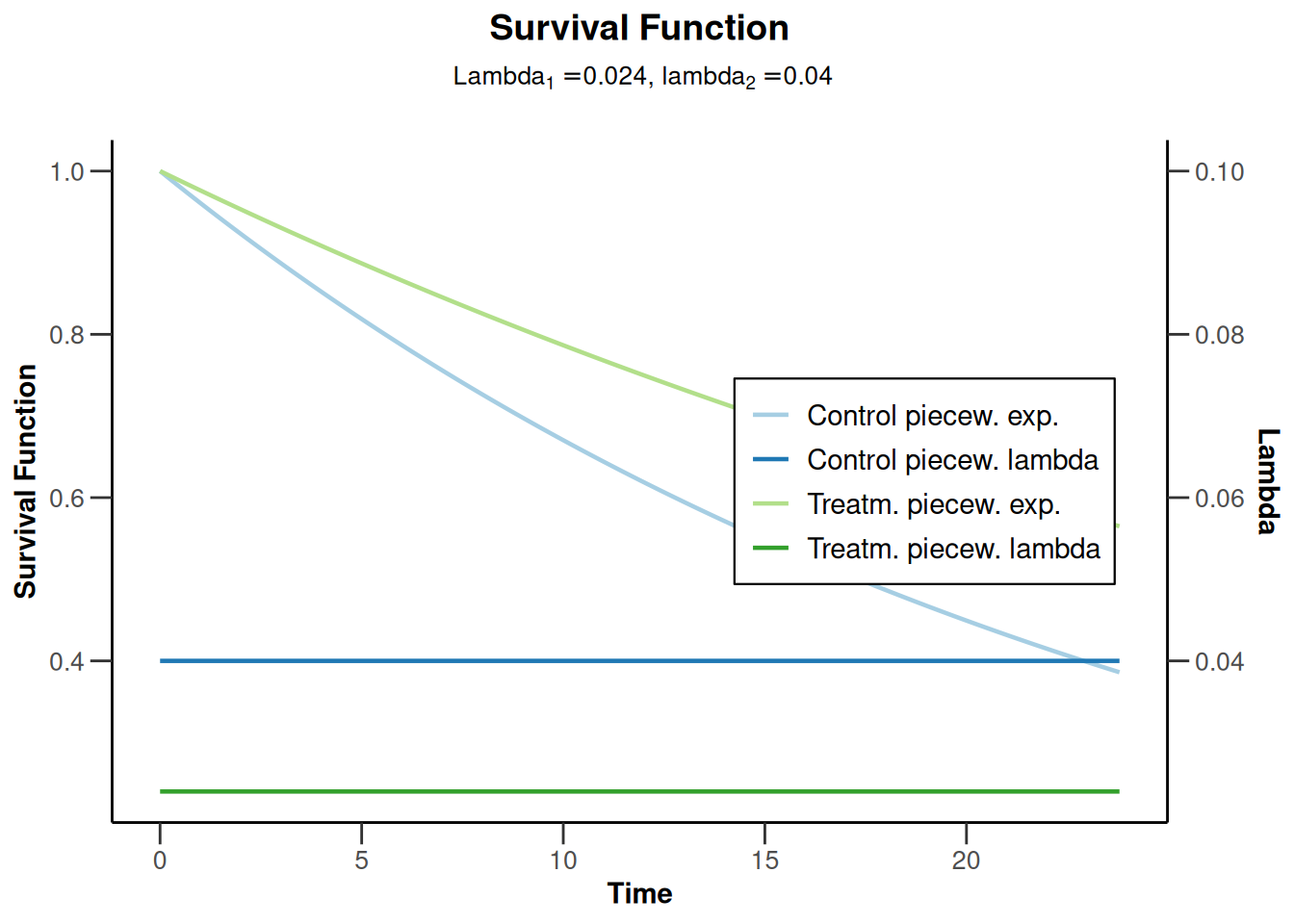
Power survival for default pi1 and pi2
powerSurvival5 <- getPowerSurvival(
maxNumberOfSubjects = 1400,
maxNumberOfEvents = 300
)
powerSurvival5 |> plot(type = 13, legendPosition = 1)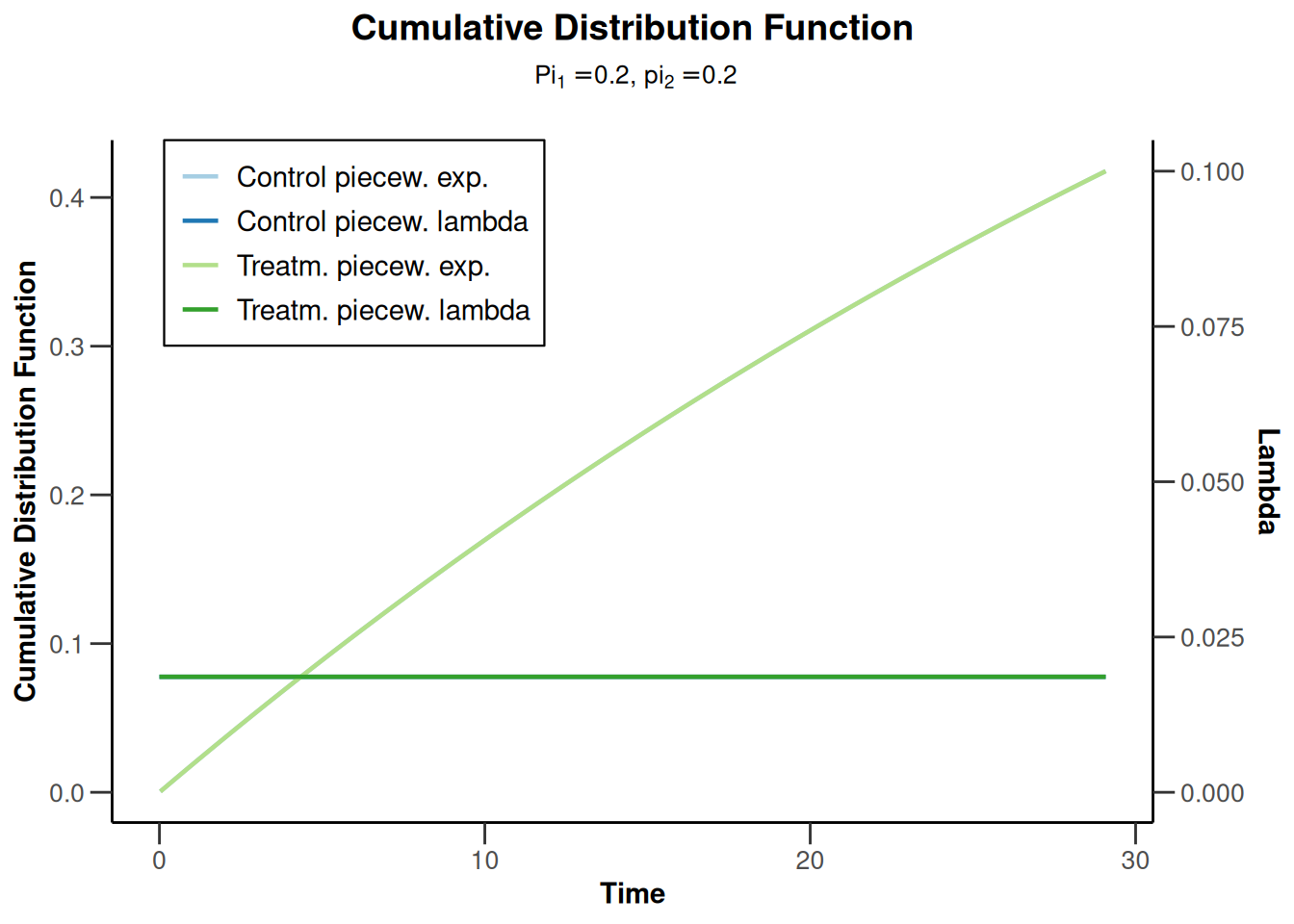
powerSurvival5 |> plot(type = 14, legendPosition = 5)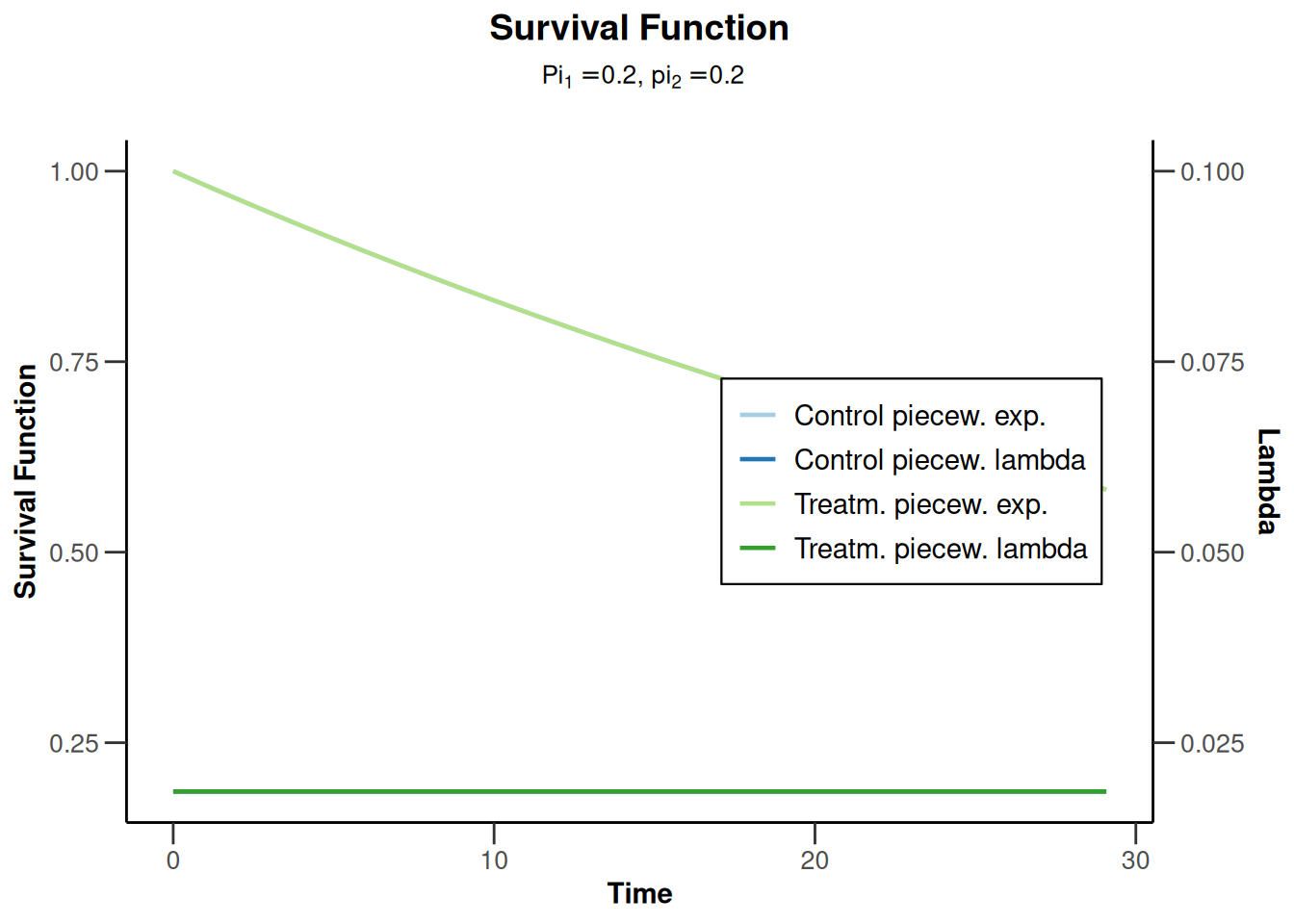
Simulation plots
Simulation means (continuous endpoint)
simulationResults1 <- getSimulationMeans(
design = getDesignFisher(kMax = 2),
plannedSubjects = c(20, 40),
maxNumberOfIterations = 1000,
seed = 12345
)
simulationResults1 |> plot(type = "all", grid = 0)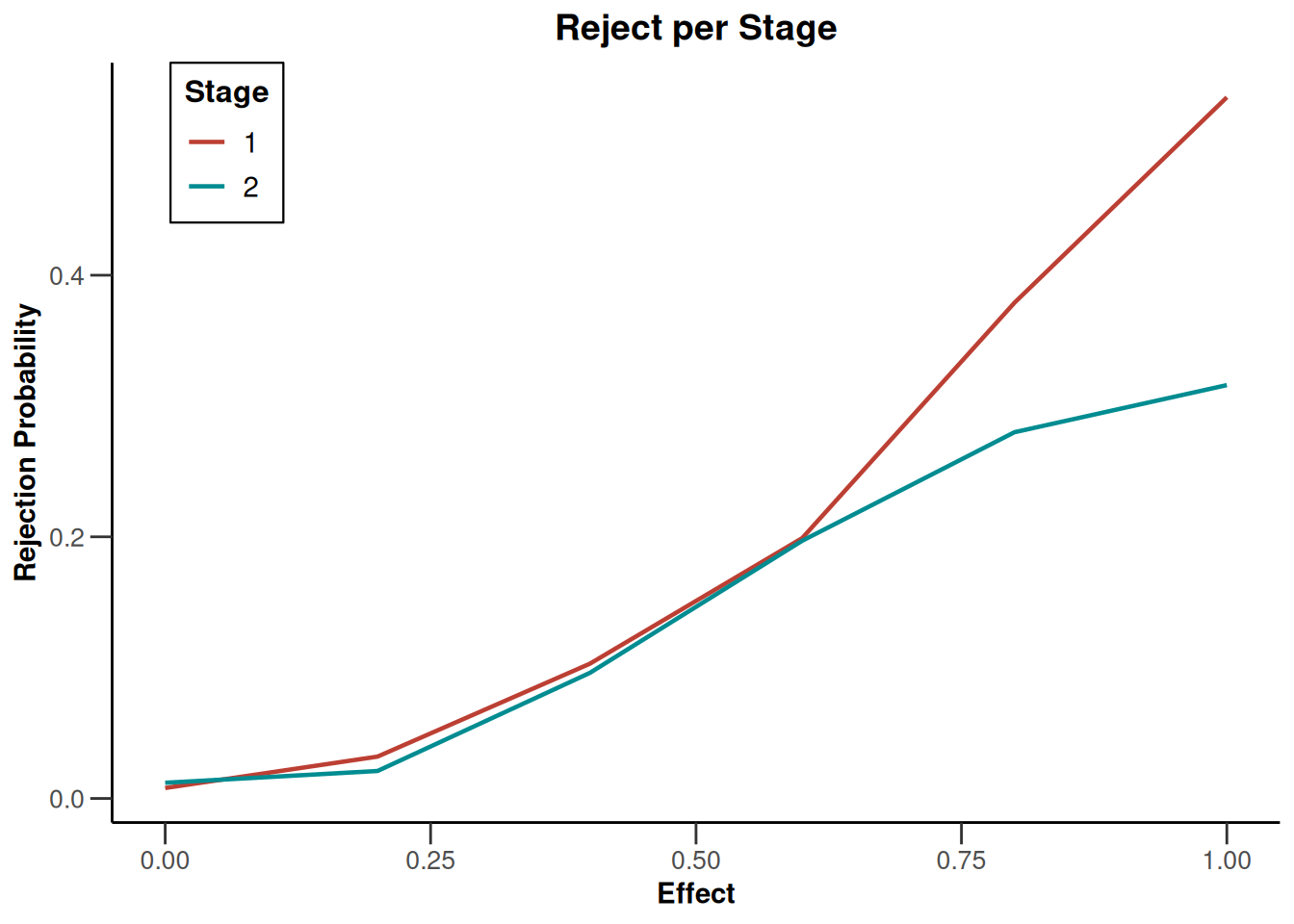
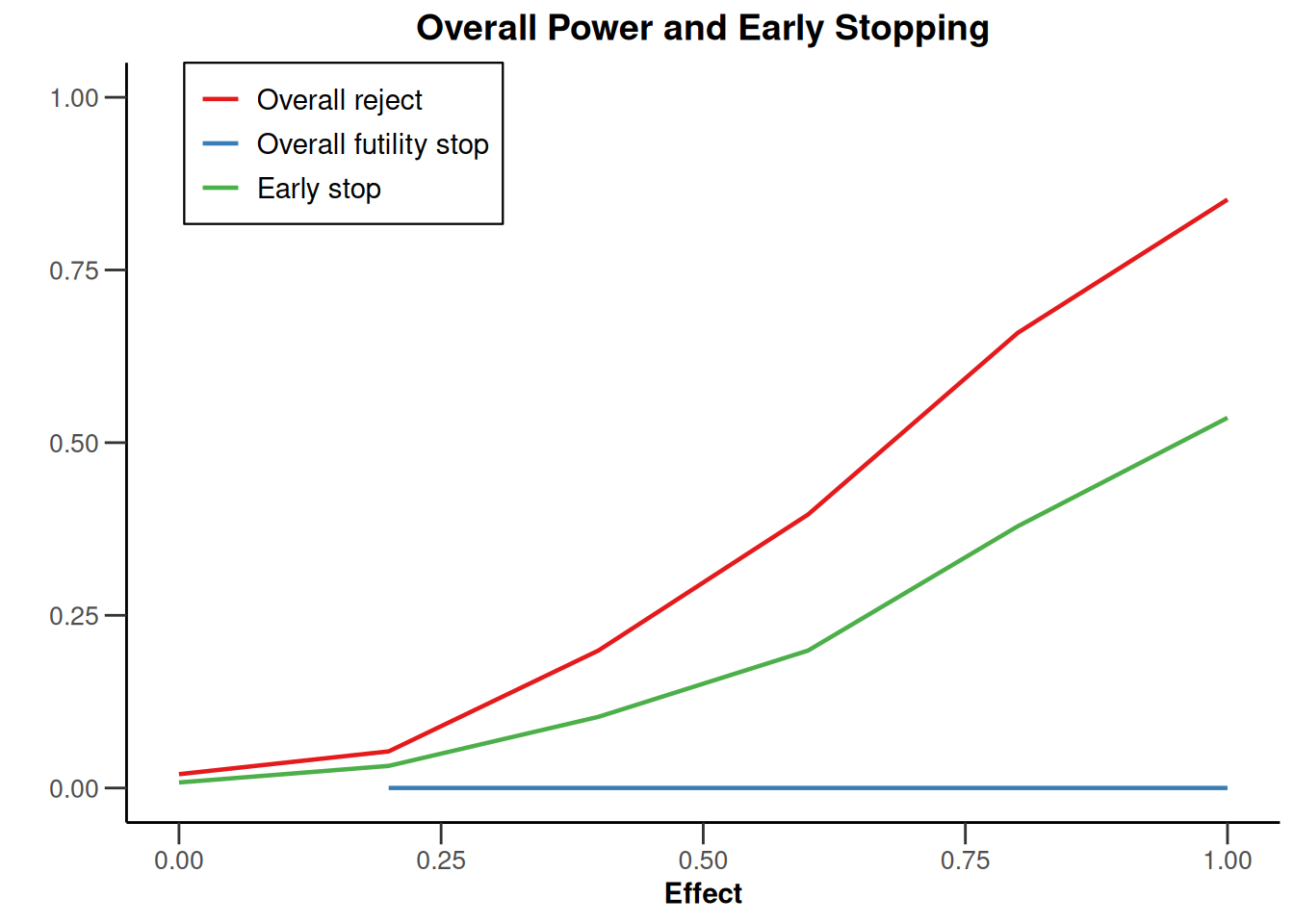
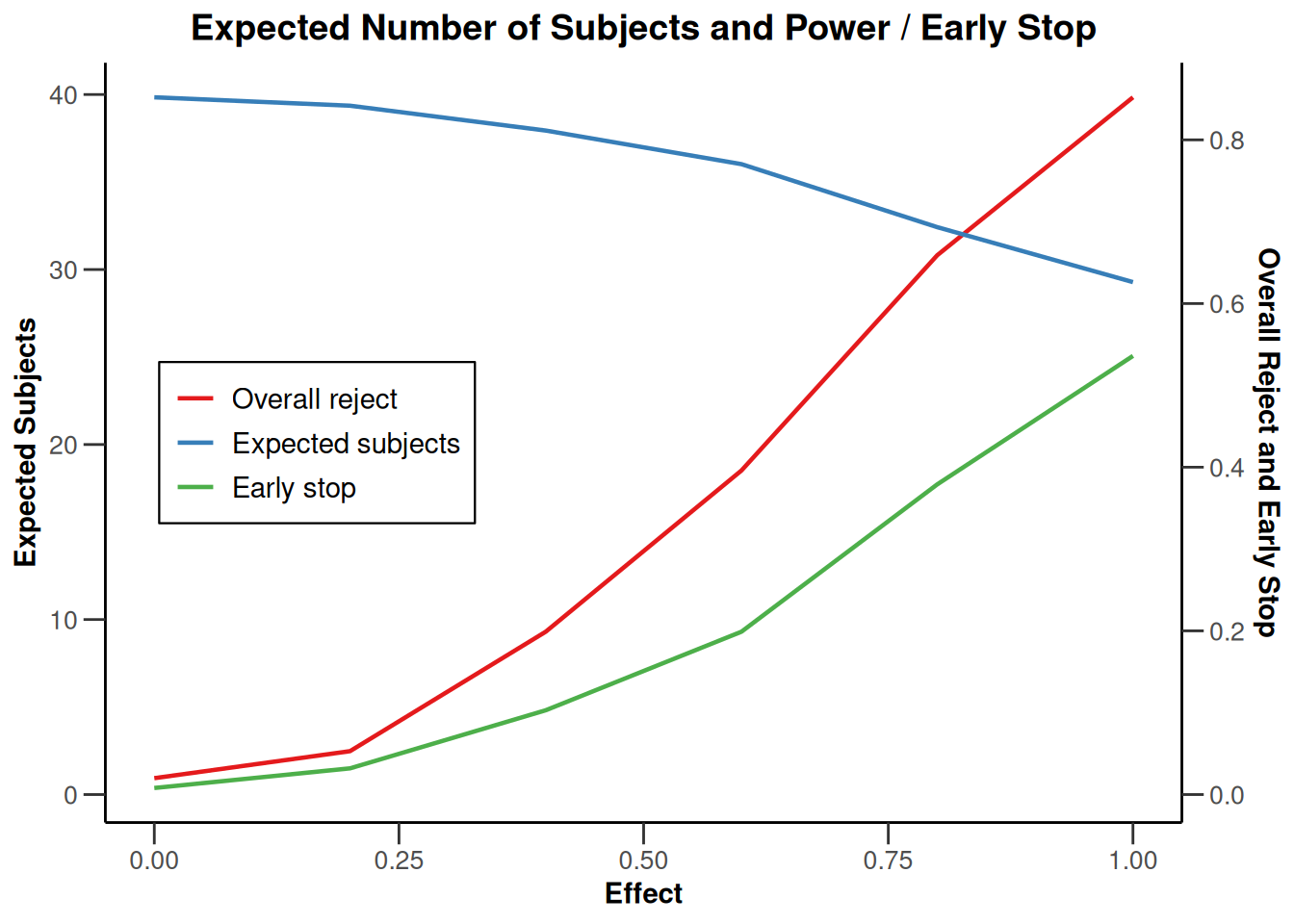
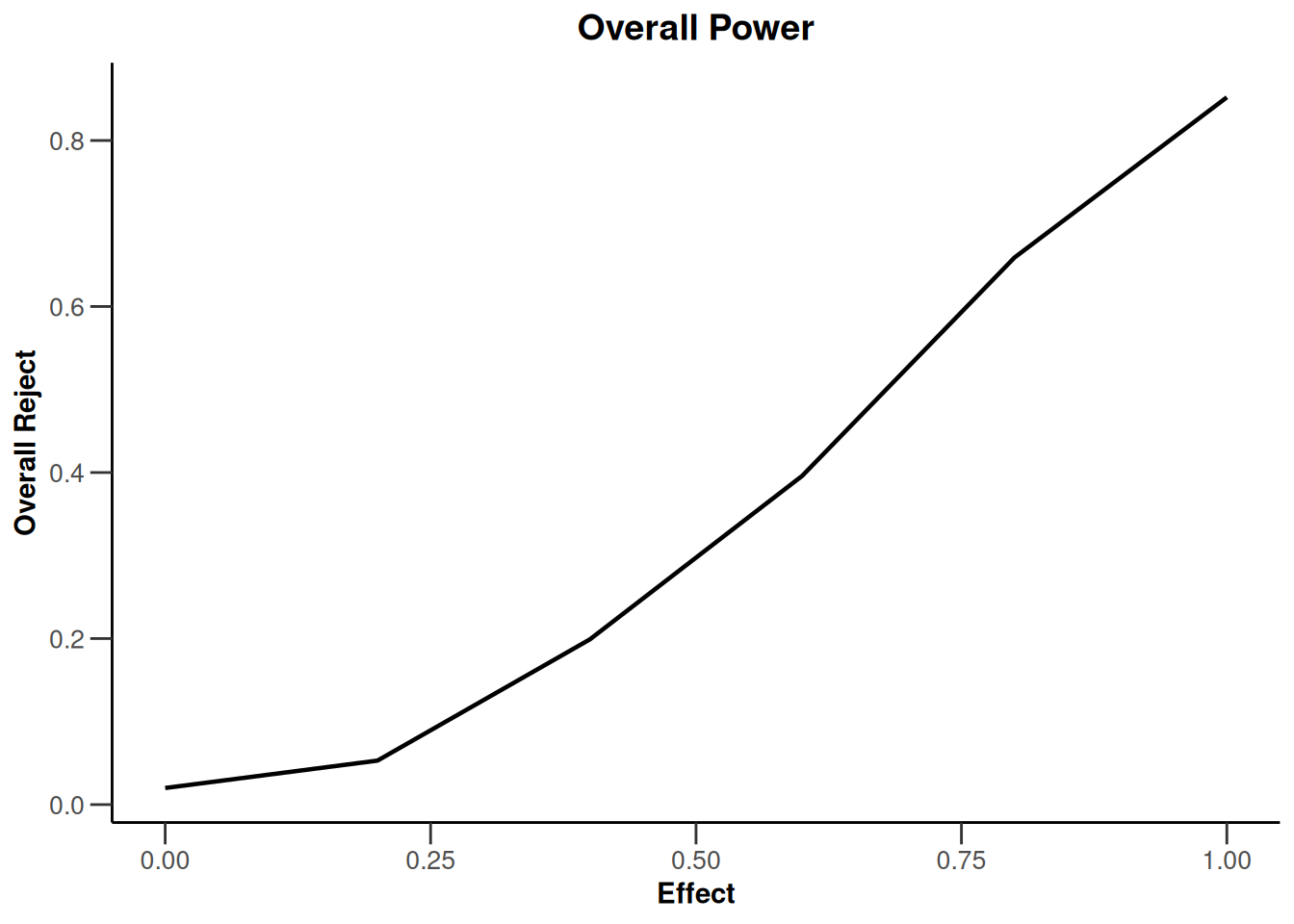
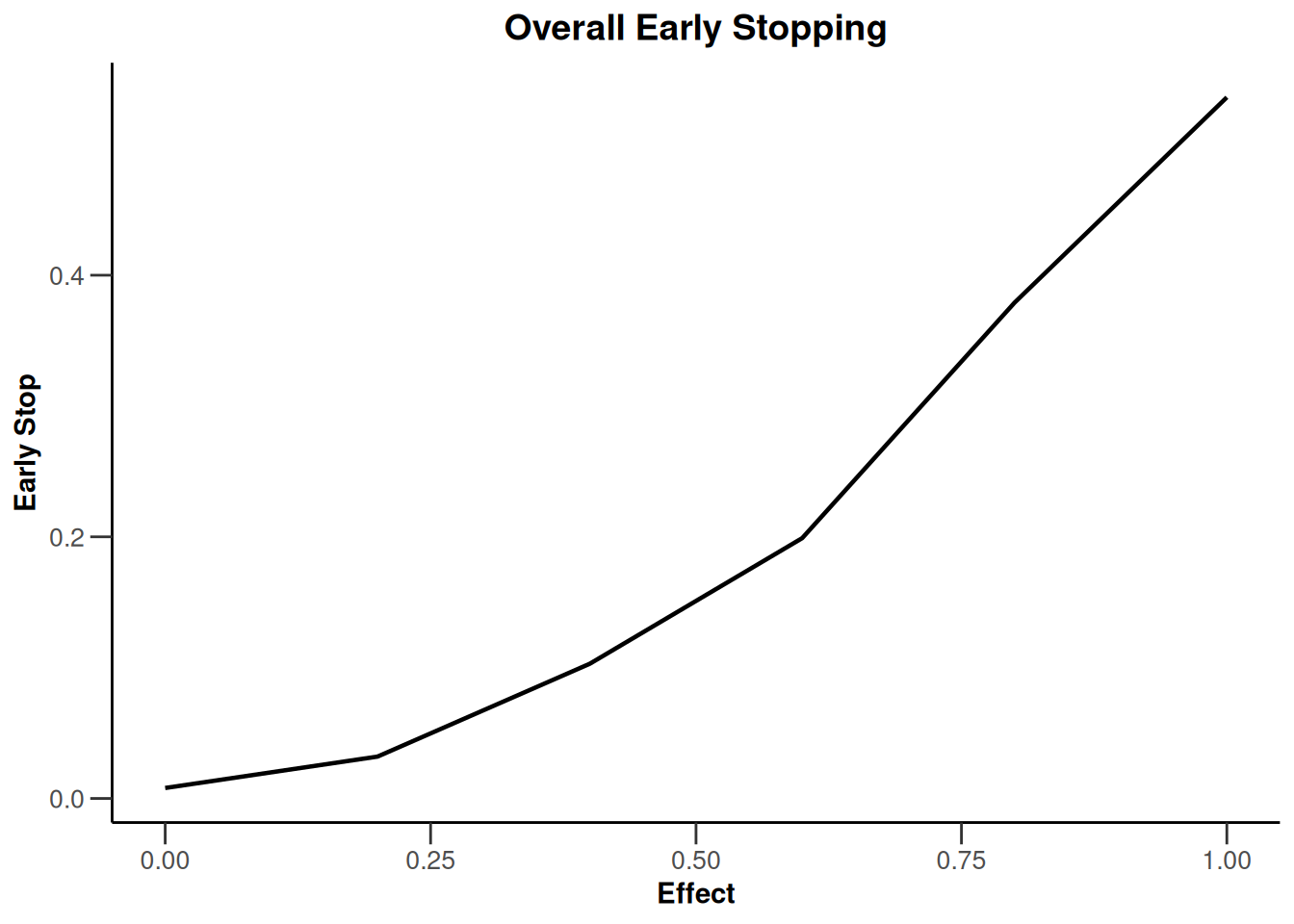
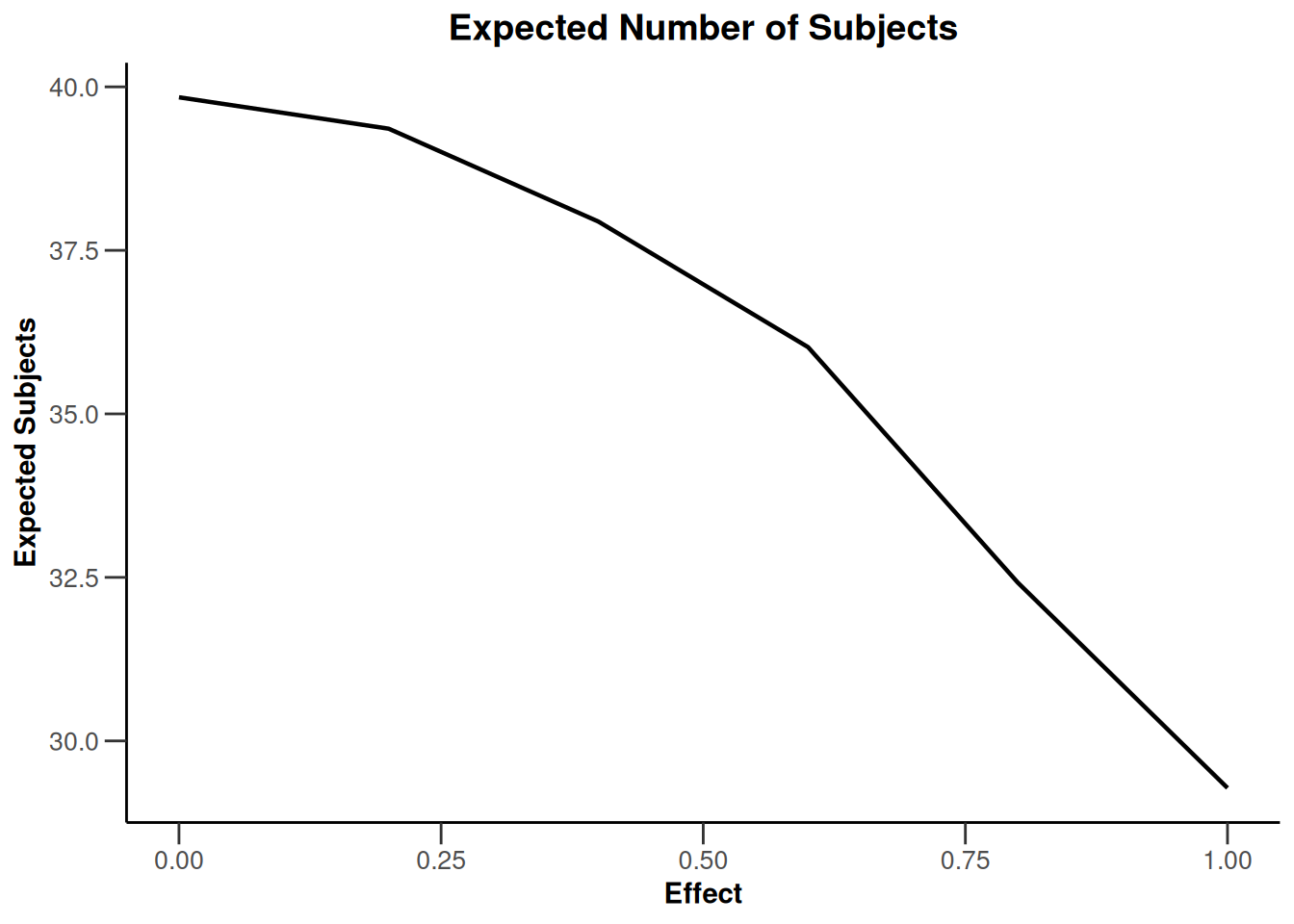
simulationResults1 |> plot(type = 4)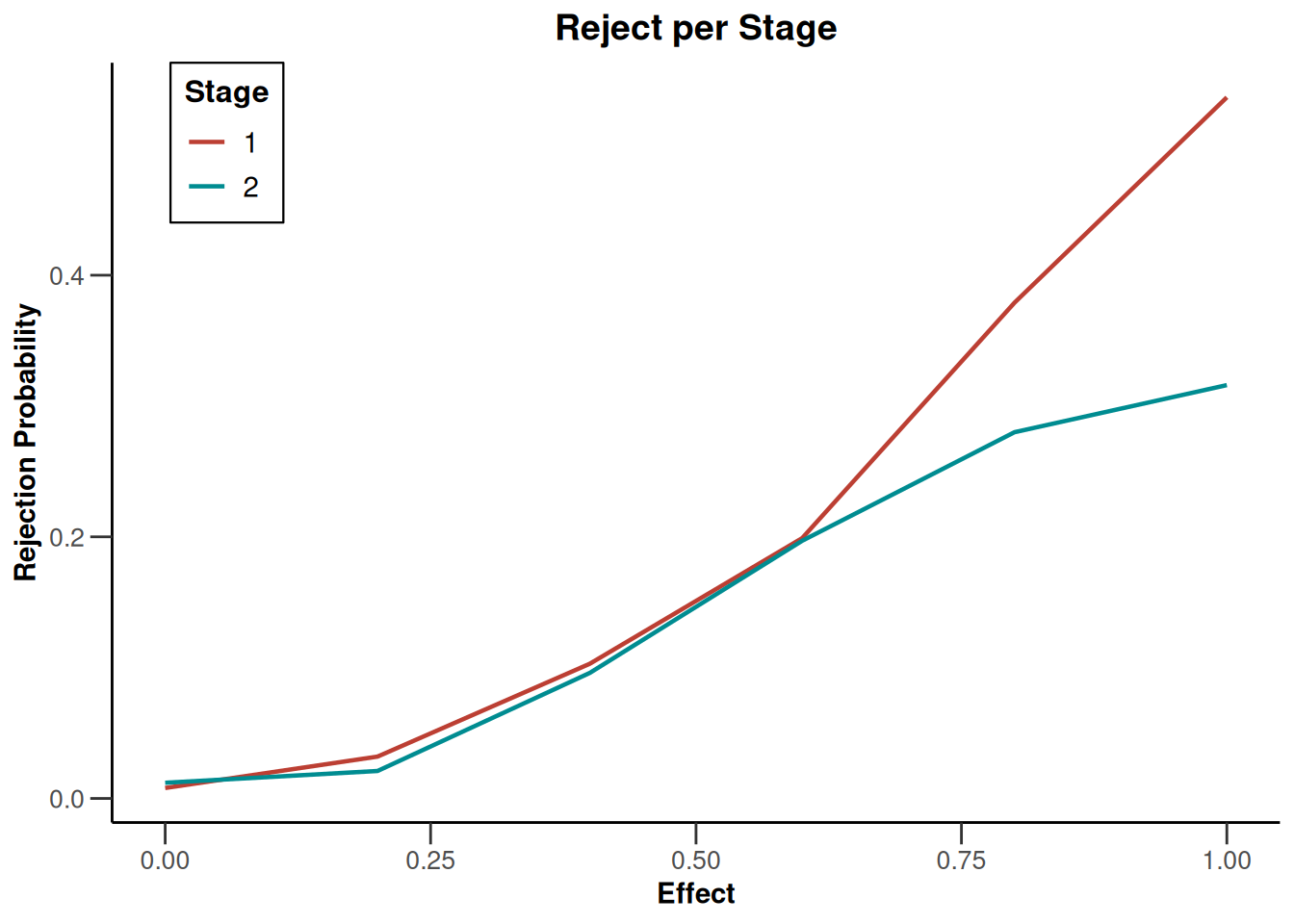
simulationResults1 |> plot(type = 5)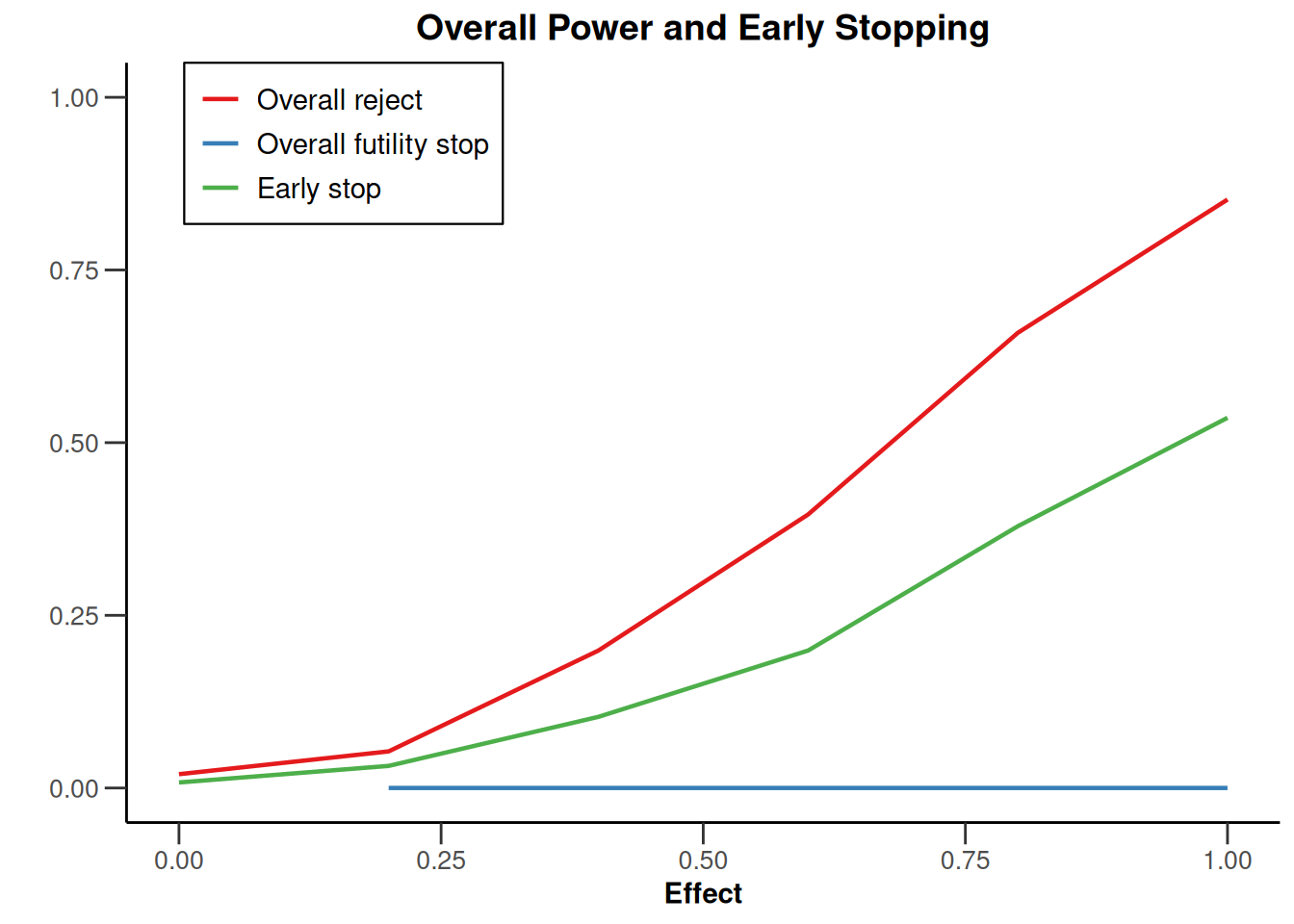
simulationResults1 |> plot(type = 6)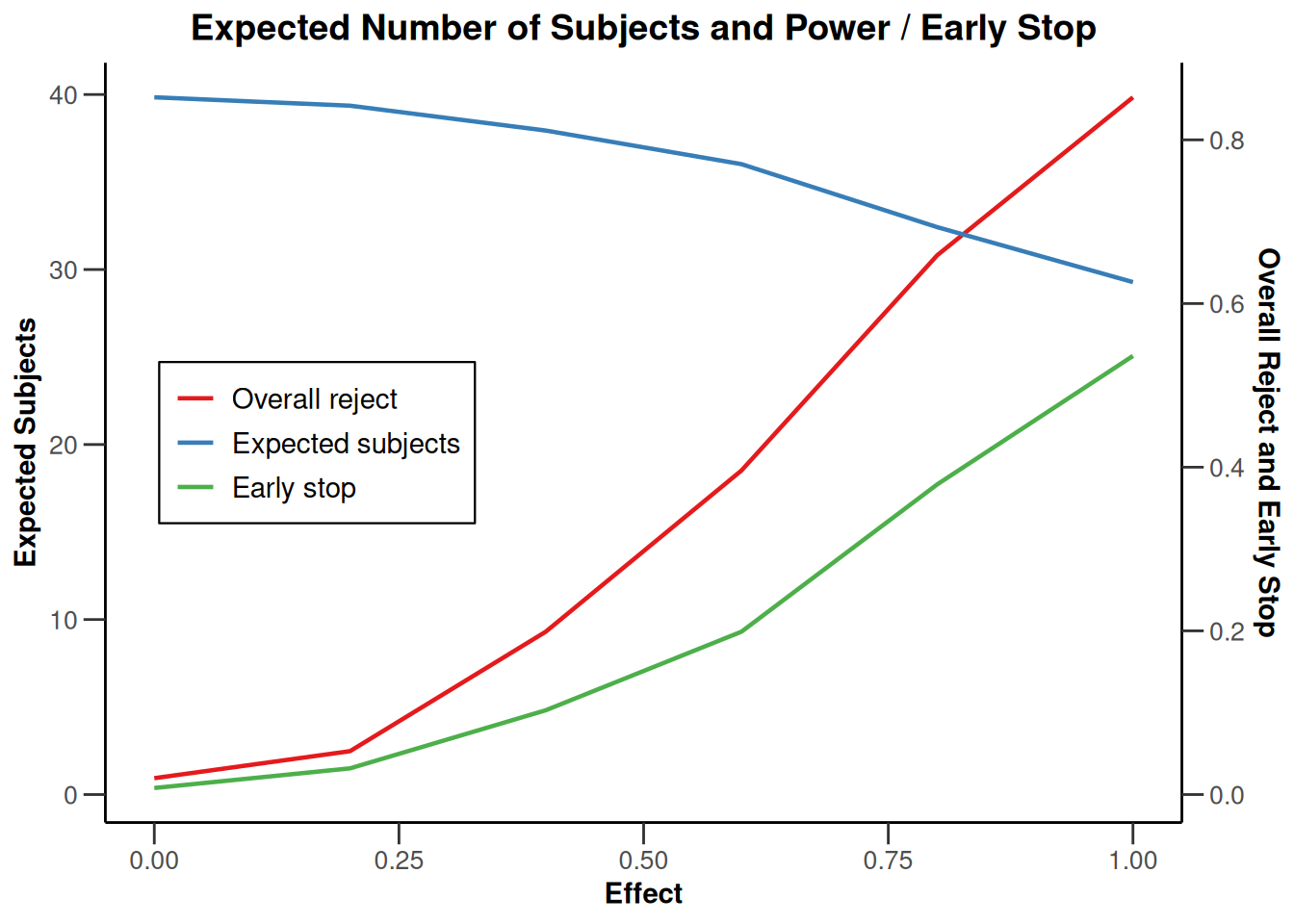
Simulation rates (binary endpoint)
simulationResults1 <- getSimulationRates(
design = getDesignFisher(kMax = 2),
plannedSubjects = c(20, 40), maxNumberOfIterations = 1000,
seed = 12345
)
plot(simulationResults1, type = 4)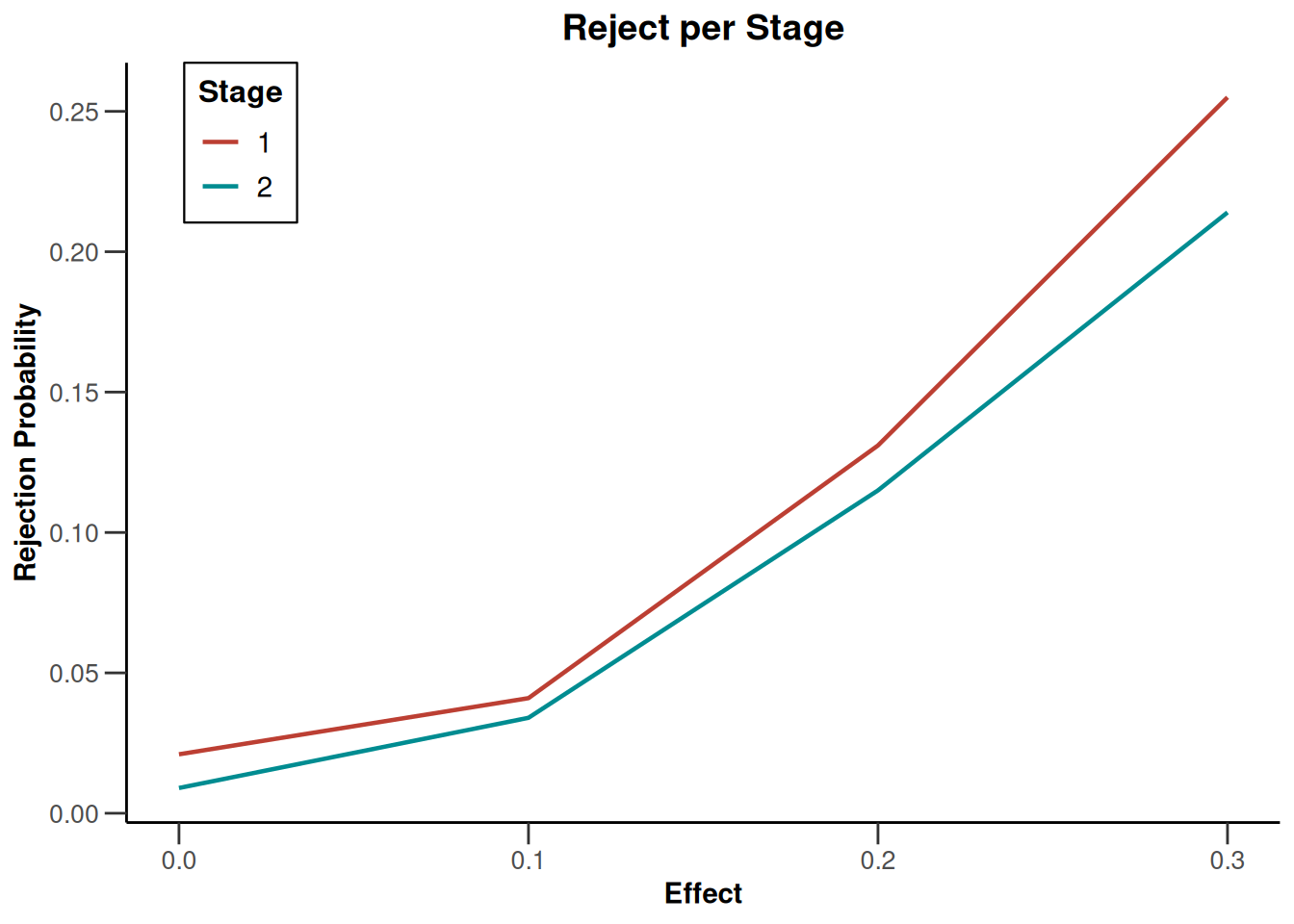
plot(simulationResults1, type = 5)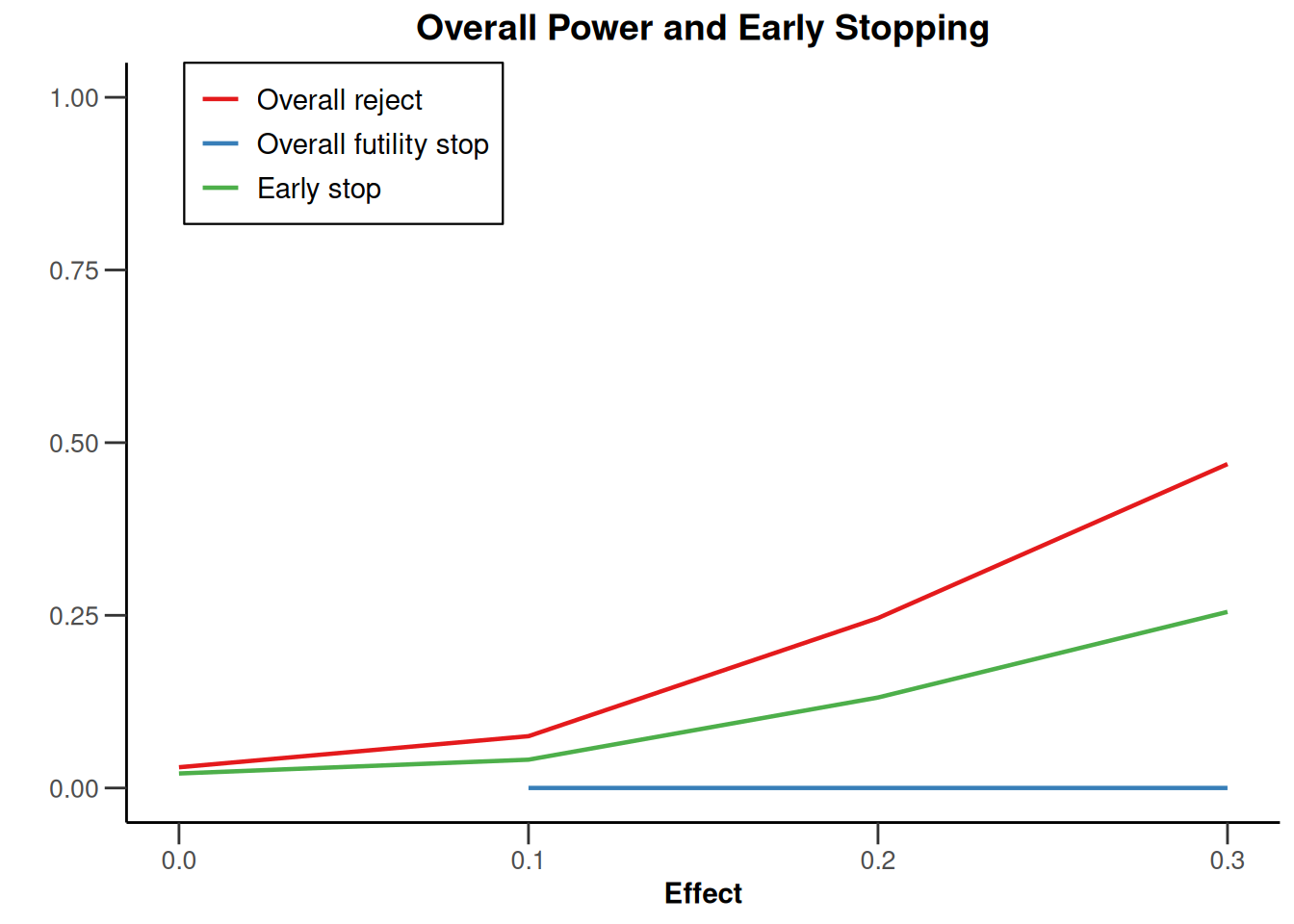
plot(simulationResults1, type = 6)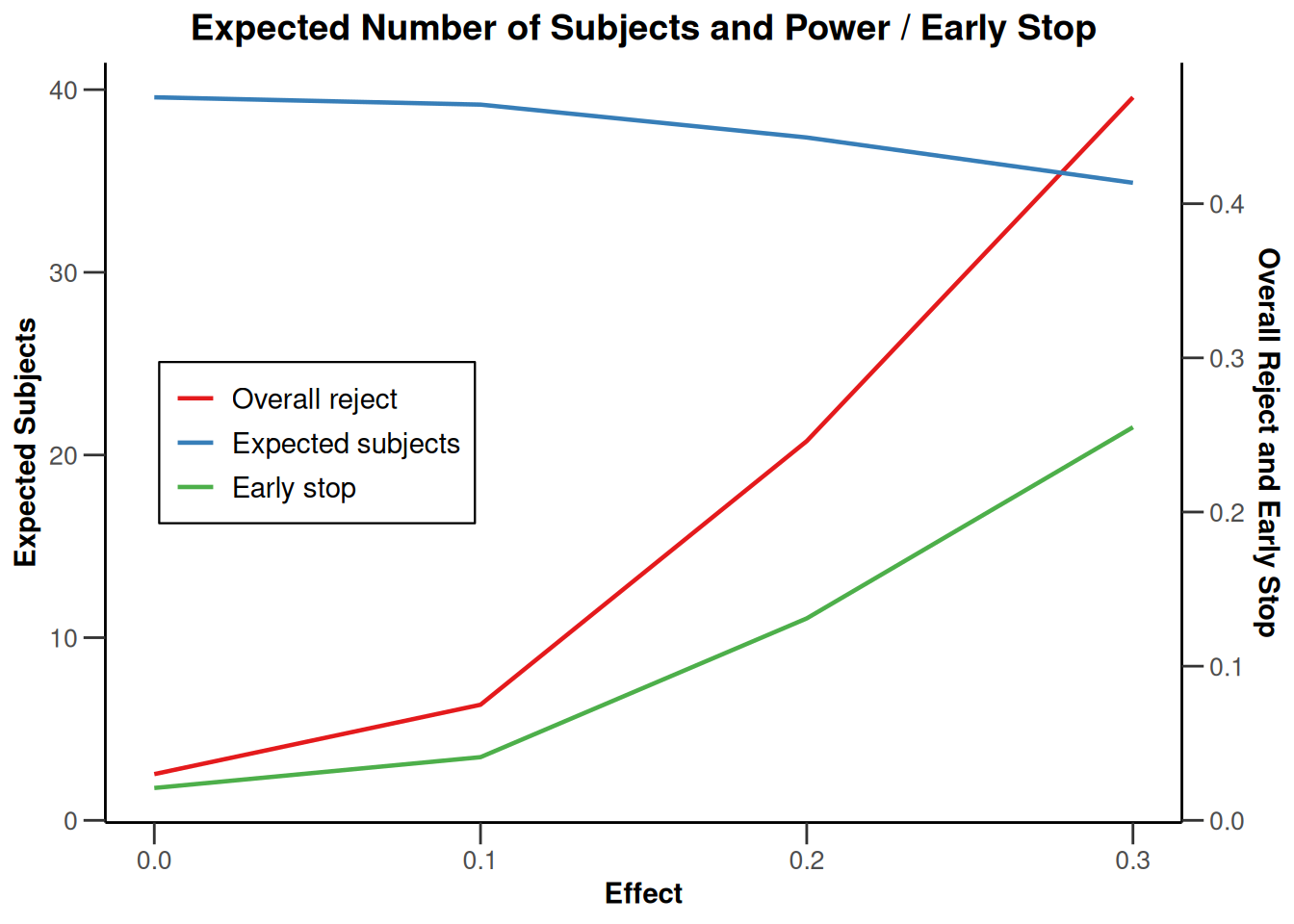
Simulation plots survival (survival endpoint)
simulationResults1 <- getSimulationSurvival(
accrualTime = 12,
maxNumberOfSubjects = 1405,
plannedEvents = 300,
maxNumberOfIterations = 1000,
seed = 12345
)
simulationResults1 |> plot(type = 5)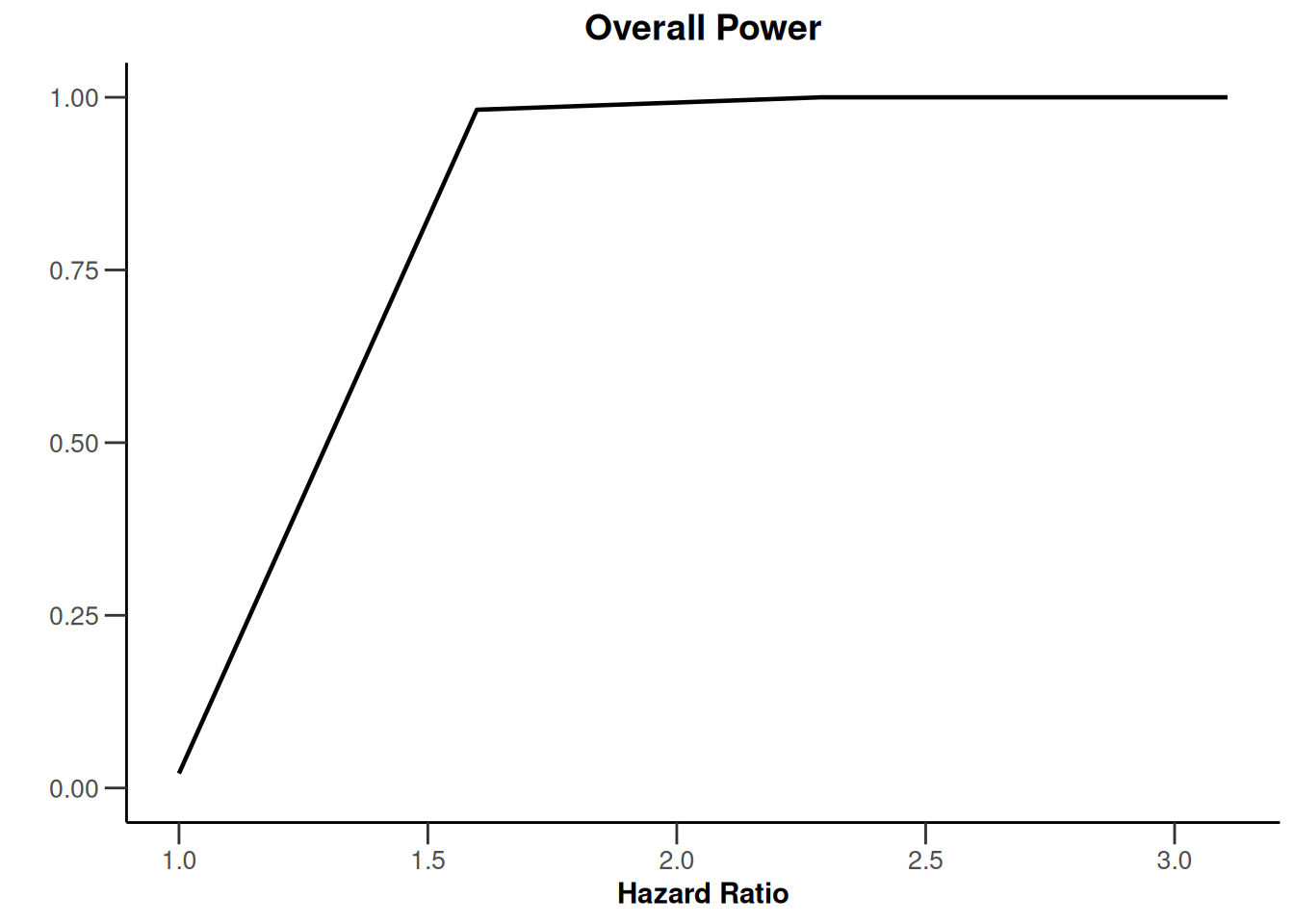
simulationResults1 |> plot(type = 6)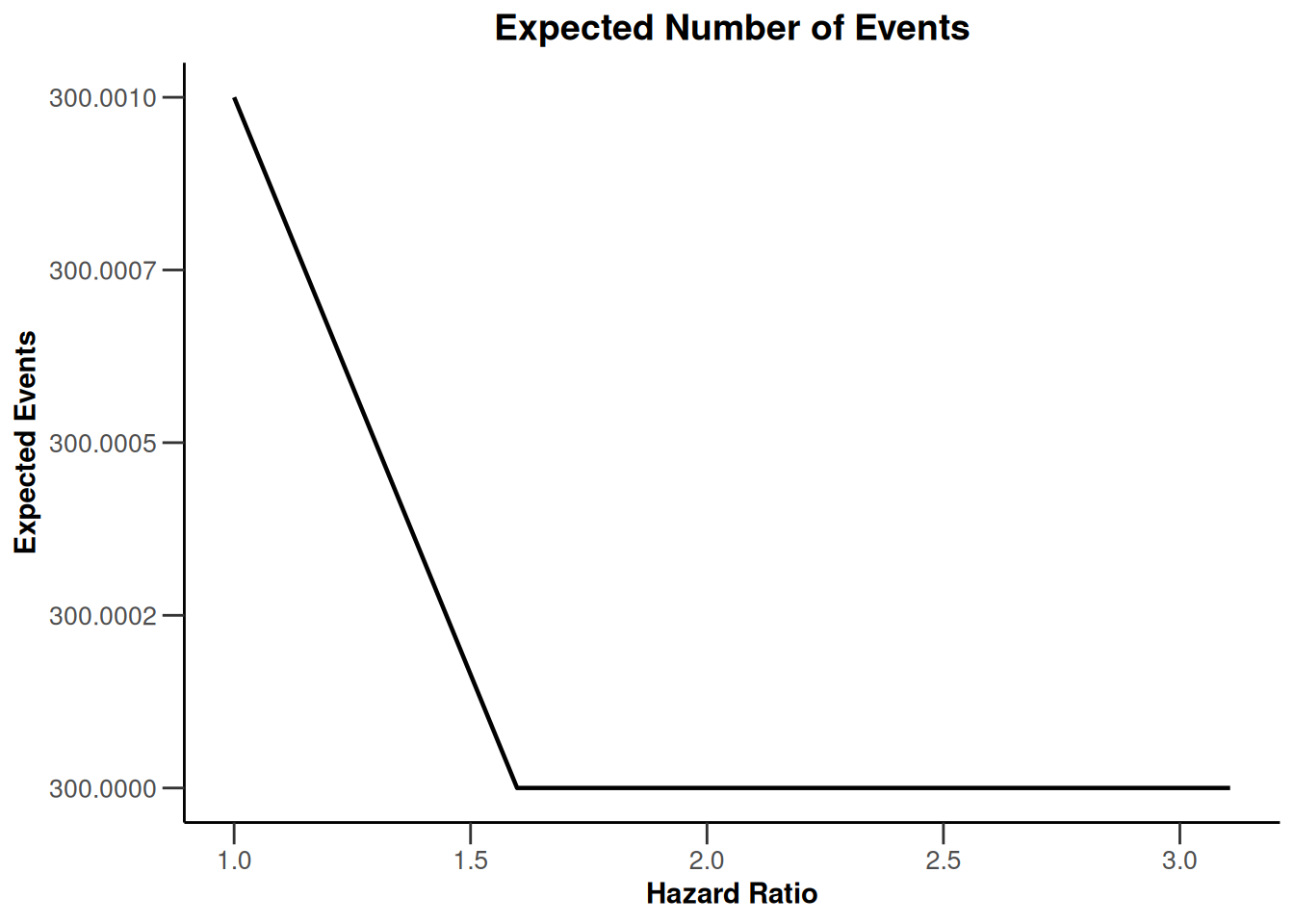
simulationResults1 |> plot(type = 7)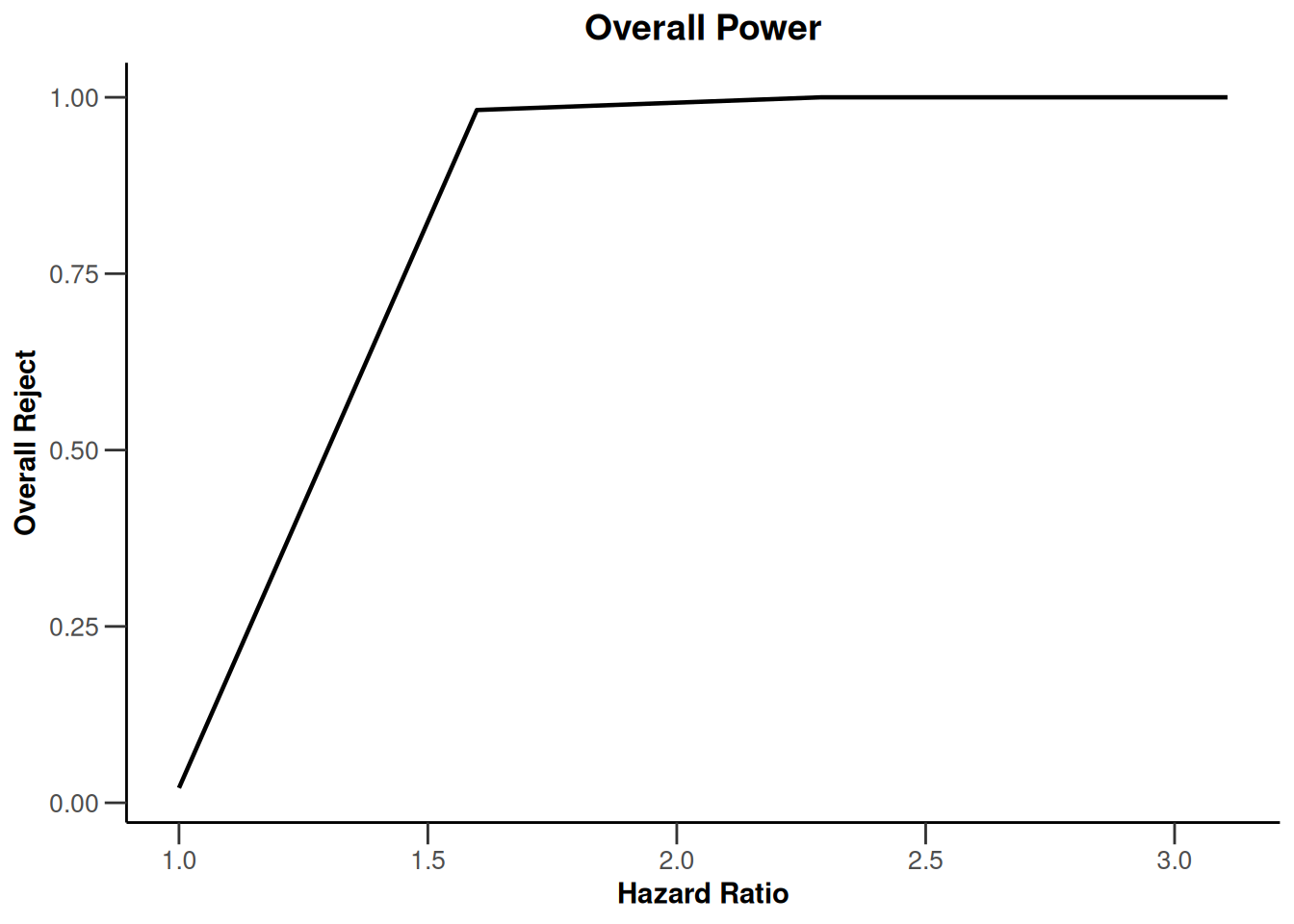
simulationResults1 |> plot(type = 9)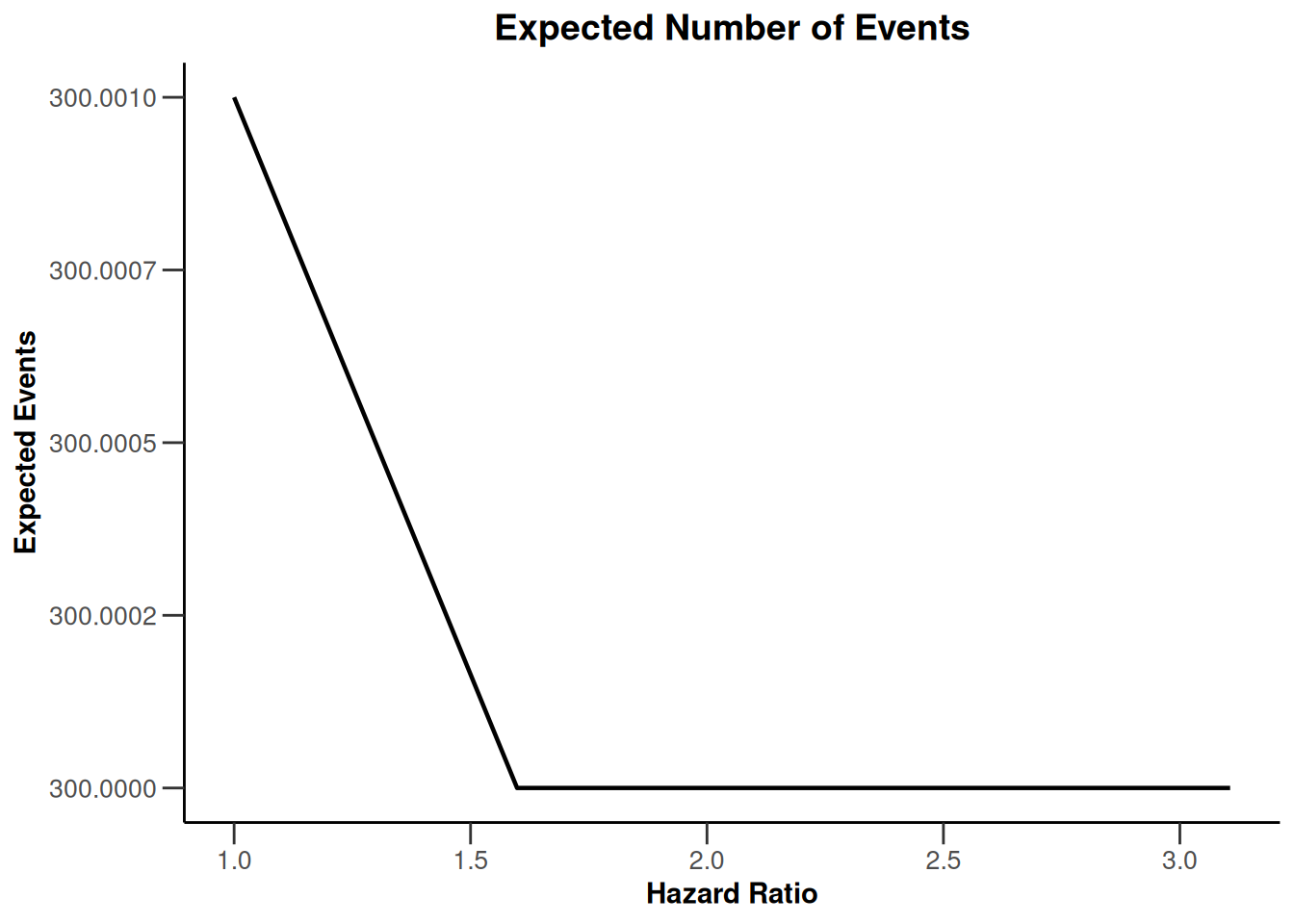
simulationResults1 |> plot(type = 10)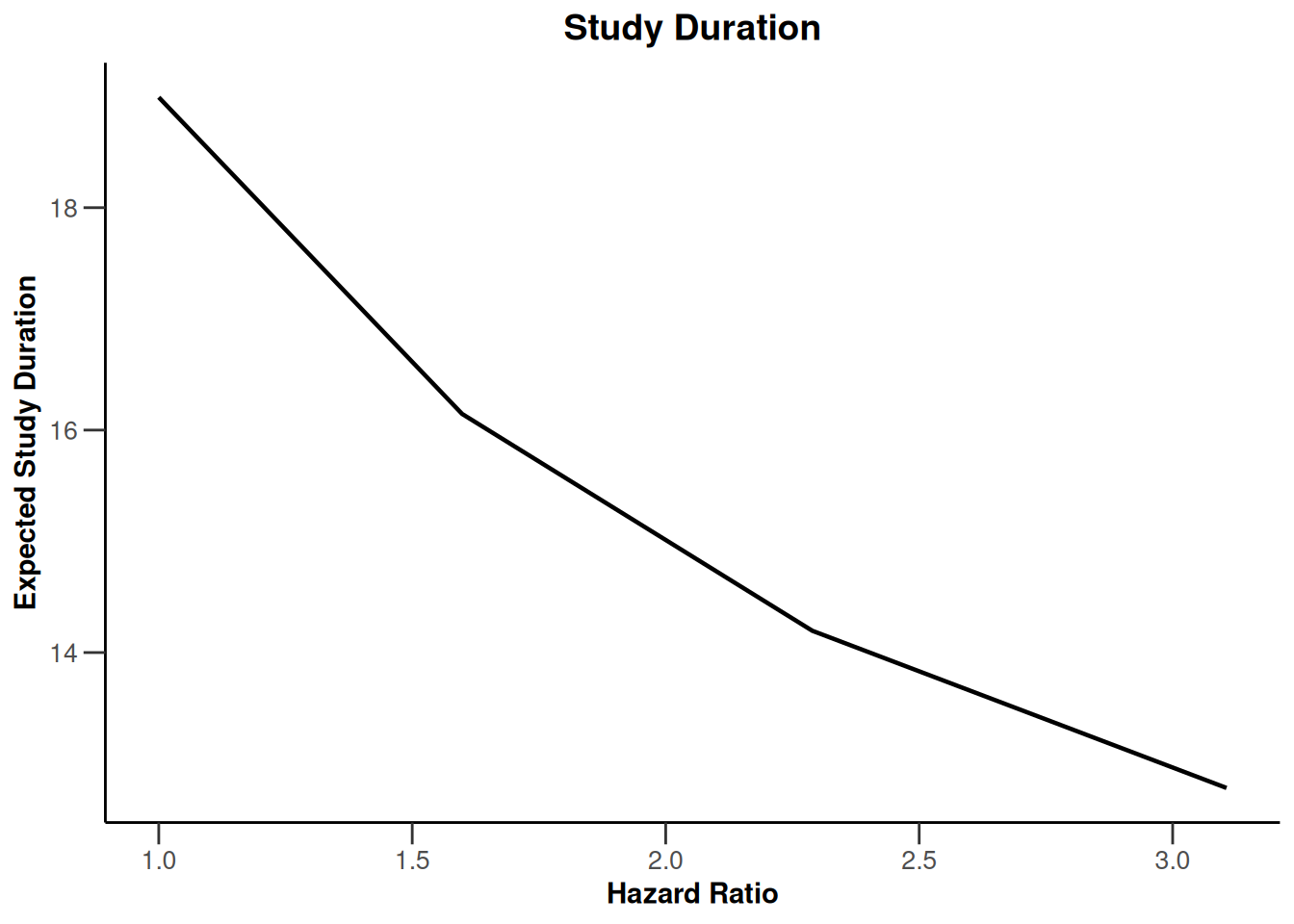
simulationResults1 |> plot(type = 11)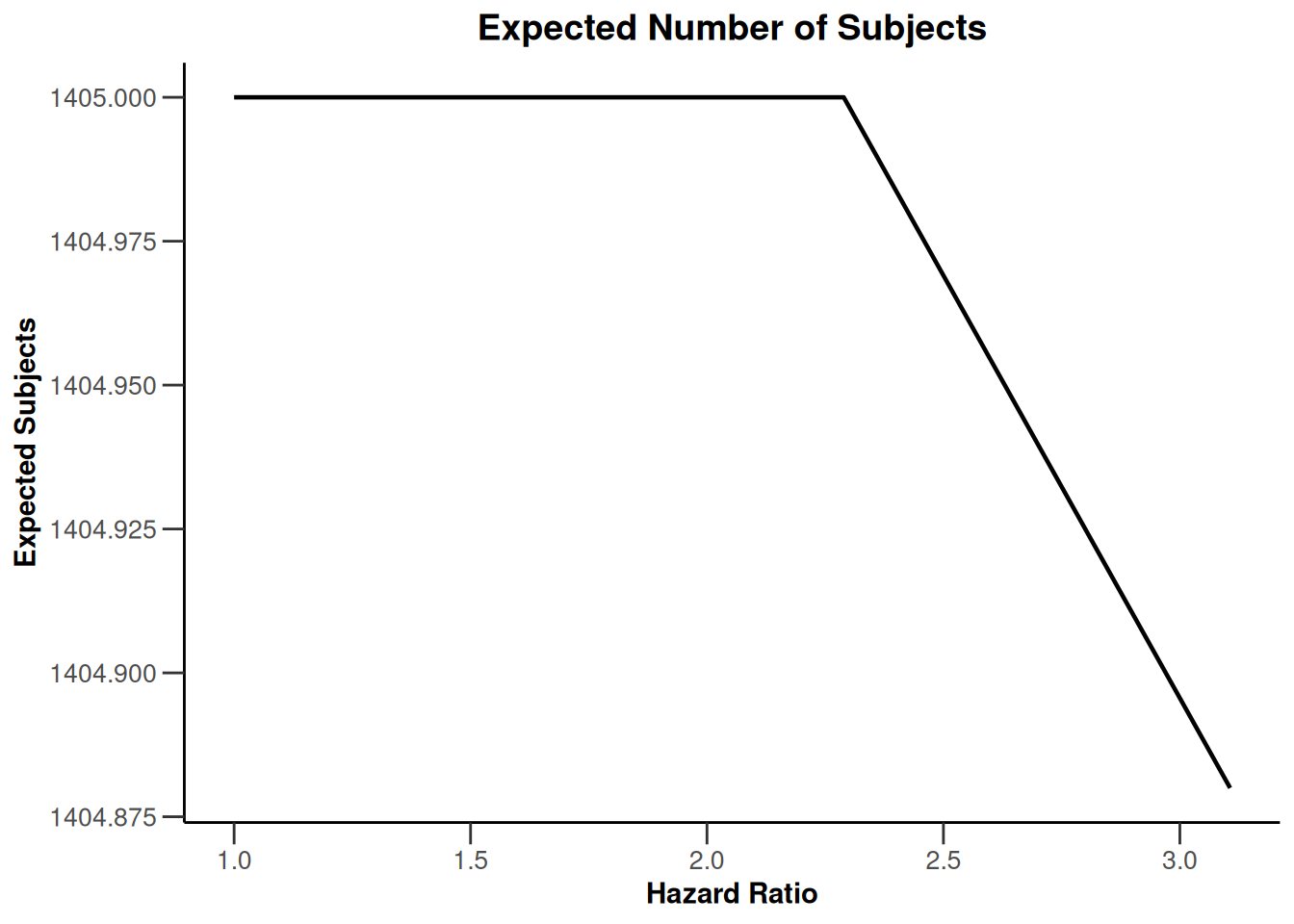
simulationResults1 |> plot(type = 12)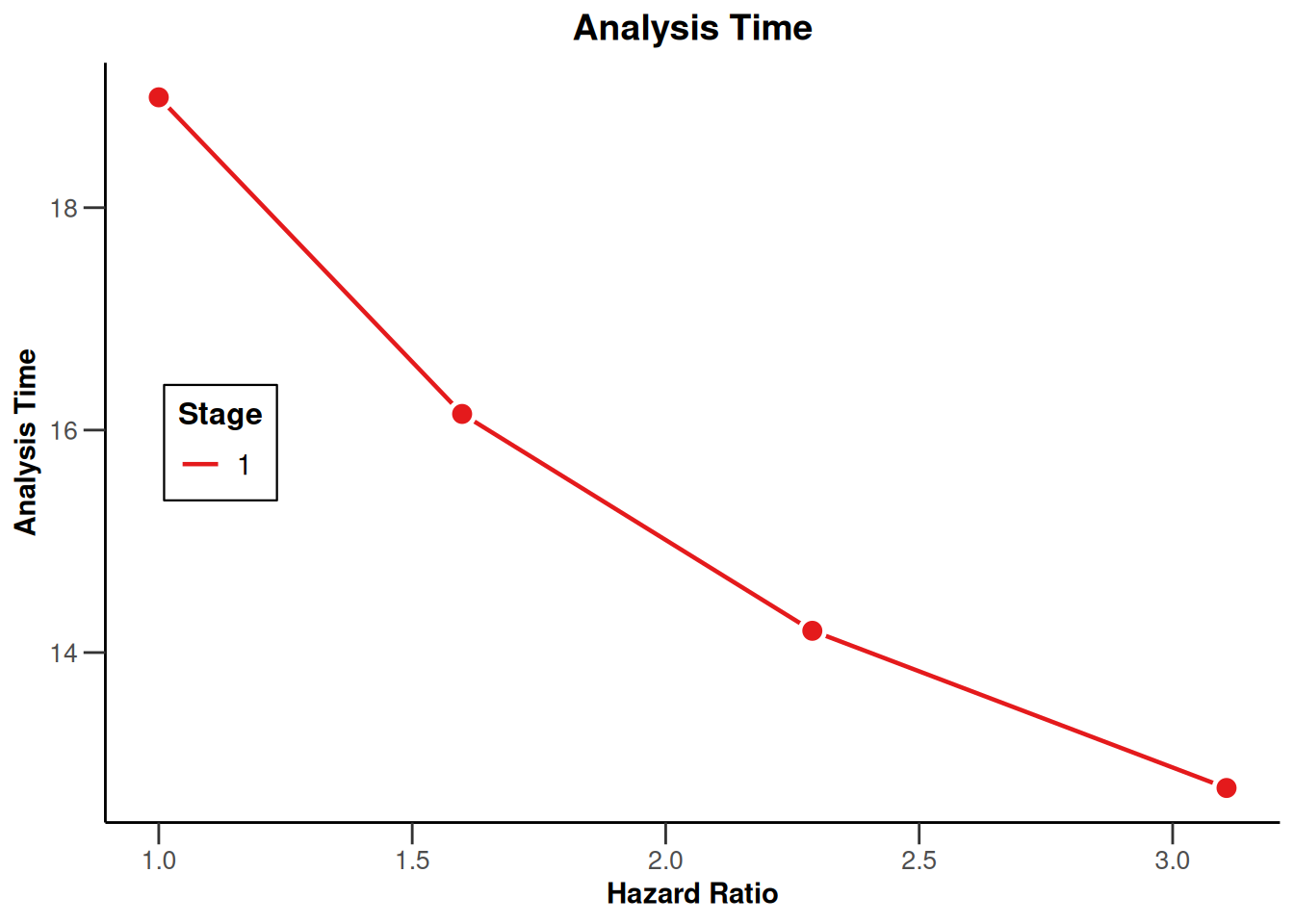
simulationResults1 |> plot(type = 13)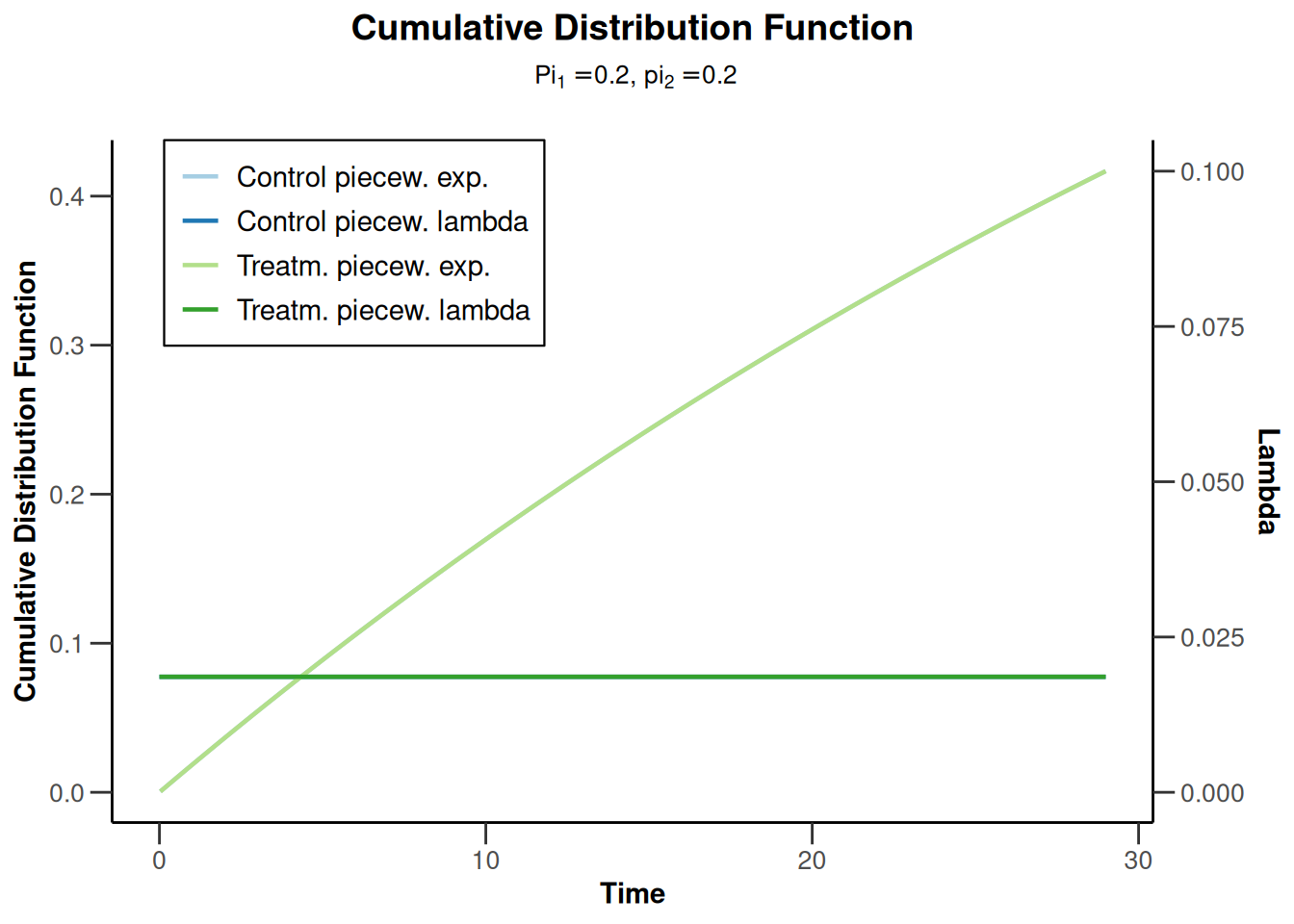
simulationResults1 |> plot(type = 14)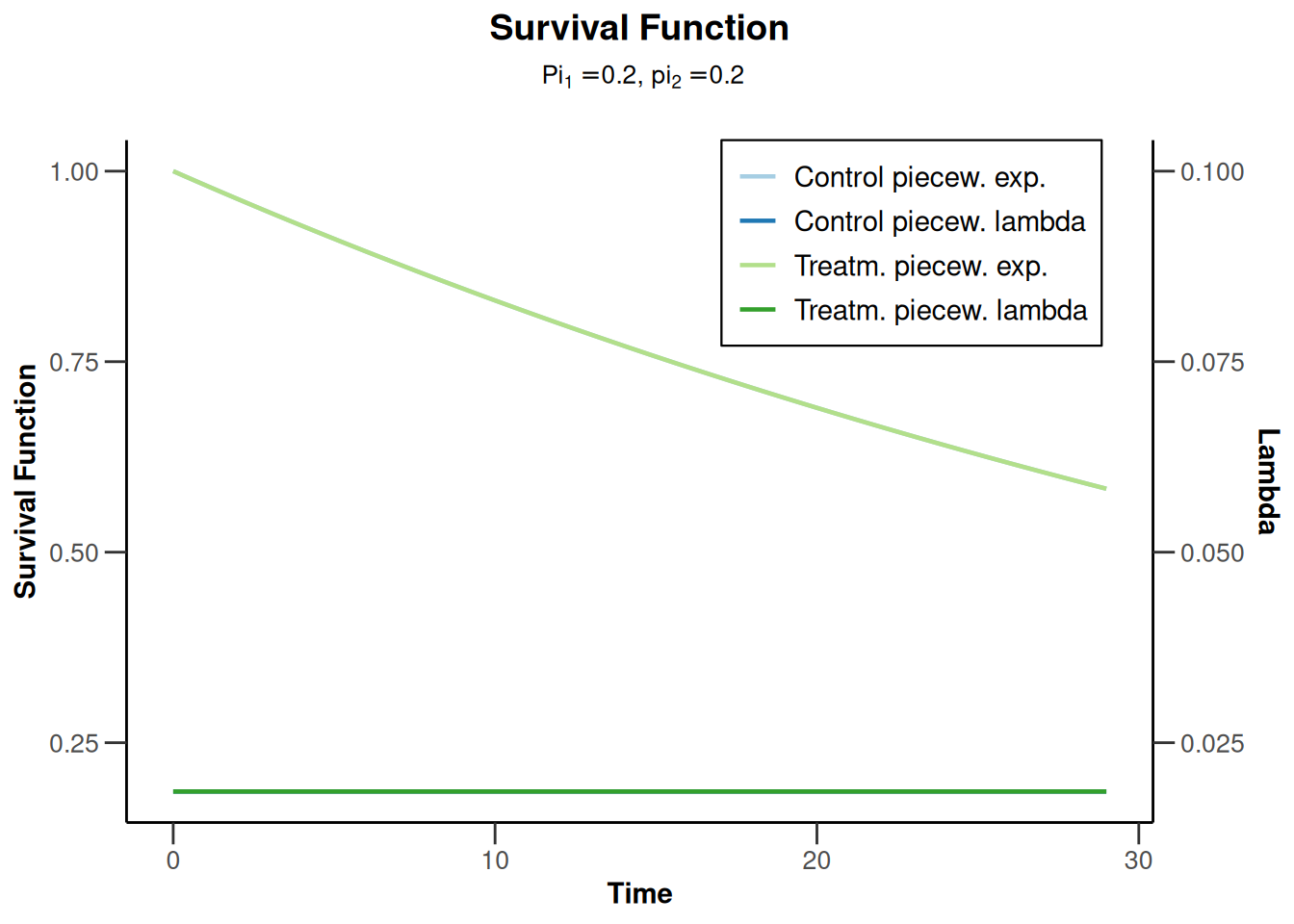
simulationResults2 <- getSimulationSurvival(
accrualTime = 12,
lambda2 = 0.03,
hazardRatio = 0.8,
maxNumberOfSubjects = 1405,
plannedEvents = 300,
maxNumberOfIterations = 1000,
seed = 23456
)
simulationResults2 |> plot(type = 13)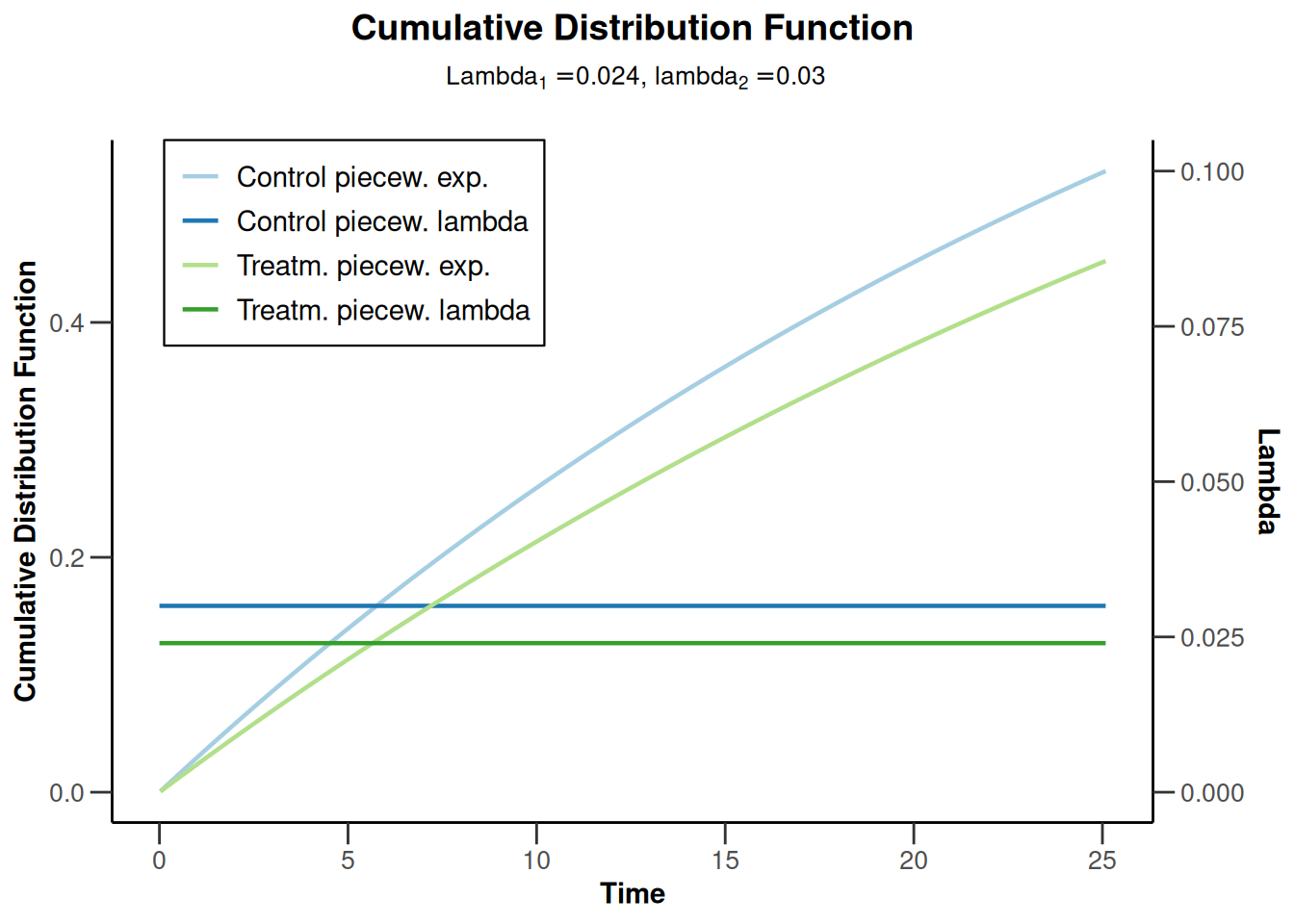
simulationResults2 |> plot(type = 14)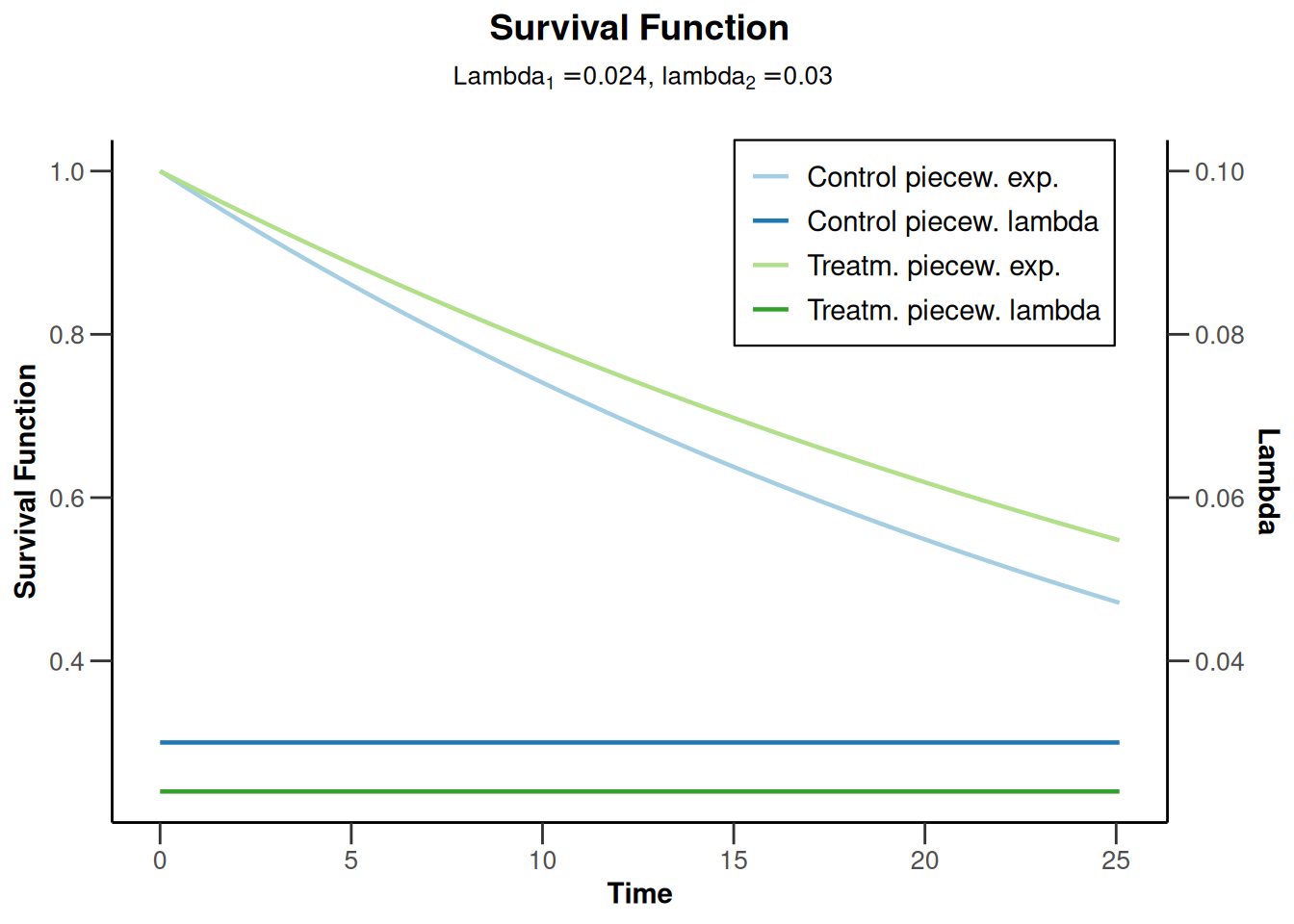
System: rpact 4.3.0, R version 4.5.2 (2025-10-31), platform: x86_64-pc-linux-gnu
To cite R in publications use:
R Core Team (2025). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
To cite package ‘rpact’ in publications use:
Wassmer G, Pahlke F (2025). rpact: Confirmatory Adaptive Clinical Trial Design and Analysis. doi:10.32614/CRAN.package.rpact https://doi.org/10.32614/CRAN.package.rpact, R package version 4.2.0, https://cran.r-project.org/package=rpact.